Abstract
The complete mitochondrial genome of Polypedilum nubifer was sequenced and annotated, and its general features and base composition were analyzed. The mitogenome was 15,896 bp long, comprising 13 protein-coding genes, 22 transport RNA genes, 2 ribosomal RNA genes, and 1 control region. The phylogenetic relationships based on the concatenated nucleotide sequences of 17 metagenomes from families Chironomidae, Ceratopogonidae, and Simuliidae were reconstructed. According to the phylogenetic topologies, P. nubifer was closely related to (P. unifascium + P. vanderplanki) in the maximum likelihood tree.
Polypedilum nubifer (Skuse Citation1889), belonging to the subfamily Chironominae, is a widely distributed and recorded species in Australia, the Federated States of Micronesia, Asia, Europe, Africa, Hawaii, and Florida. In China, the species has been found in the Palearctic (Liaoning, Ningxia, Inner Mongolia, Tianjin, Henan, Shananxi, Gansu, Anhui, and Xinjiang) and Oriental (Sichuan, Yunnan, Fujian, Taiwan, Guizhou, Guangdong, Guangxi, and Hainan) regions (Wang et al. Citation2020). Most larvae of the genus Polypedilum grow in contaminated water. Thus, P. nubifer can be used as an indicator species for monitoring water environments (Paine and Gaufin Citation1956; Armitage et al. Citation1995). Polypedilum nubifer is a common nuisance midge found in the tropical and subtropical waters in the Afrotropical, Palearctic, Oriental, Australasian, and Nearctic regions. Populations can become extremely abundant in warm, shallow, and eutrophic waters subjected to seasonal drying (CitationTabaru et al. 1987; Trayler et al. Citation1994; Cranston Citation2007; Jacobsen and Perry Citation2007; Cranston et al. Citation2013).
The specimen in this study was collected from the Wujiashan Mountains (31°0845′N, 115°8189′E), altitude 487 m, Dahechong Village, Yingshan County, Huanggang City, Hubei Province, CHINA) on 9 August 2020 using a light trap. Samples were collected by Yue Fu (email: [email protected]) and deposited in the Biodiversity Herbarium of Huanggang Normal University (http://shengwu.hgnu.edu.cn/2018/1130/c435a7076/page.htm, Yue Fu) under the voucher number HGNU-Ydbs152. Whole genomic DNA was extracted using the DNeasy Animal Tissue Kit (Tiangen, China), and the mitochondrial genome was sequenced using the Illumina HiSeq X System. Reads were assembled using NOVOPlasty (Dierckxsens et al. Citation2016), and the mitochondrial genome was assembled using SPAdes (version v3.11.1; Bankevich et al. Citation2012). Phylogenetic analyses were reconstructed using PhyloSuite (Zhang et al. Citation2020) with several plug-in programs including MAFFT using the ‘–auto’ strategy for multiple sequence alignment (Katoh and Standley Citation2013). The AICc criterion in PartitionFinder2 was used to select best-fit partitioning schemes and models (Lanfear et al. Citation2017). The phylogenetic tree was inferred using IQ-TREE (Minh et al. Citation2013; Nguyen et al. Citation2015) and edited in iTOL (Letunic and Bork Citation2019).
The complete mitogenome of P. nubifer was 15,896 bp long (GenBank accession number: MZ747090). It consists of 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes (totaling 37 genes), and 1 control region. The genomic nucleotide composition was 40.07% A, 37.55% T, 13.05% C, and 9.33% G. The total length of the 13 PCGs in the mitochondrial genome was 11,196 bp. Four genes had overlapping regions, with a total overlapping length of 18 bp. The two longest overlapping regions (7 bp) were located between atp8/atp6 and nad4/nad4l. There were 26 intergeneric spacers with a total length of 537 bp, ranging from 1 to 87 bp. The longest intergenic region was located between trnA and trnR. The initiation codons of PCGs complied with the ATN rule: six genes (cox2, atp6, cox3, nad4, nad4l, and cob) with an ATG start codon, two genes (atp8 and nad3) with an ATA start codon, one gene (nad2) with an ATT start codon, one gene (nad6) with an ATC start codon, and two genes (nad5 and nad1) with a GTG start codon. All PCGs had a TAA stop codon. The length of the tRNA genes ranged from 65 to 72 bp, with a total length of 1,487 bp. The lengths of the 12S rRNA and 16S rRNA were 810 and 1,430 bp, respectively.
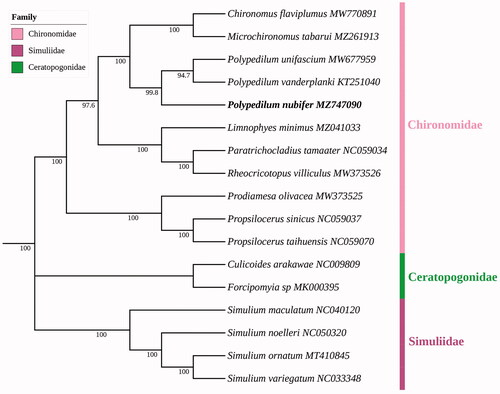
Maximum likelihood analysis showed species of the same family clustered together (). P. nubifer and (P. unifascium + P. vanderplanki) were clustered together with a bootstrap score of 99.8%; all of them belong to genus Polypedilum. Further, P. nubifer + (P. unifascium + P. vanderplanki) were closely related to Chironomus flaviplumus + Microchironomus tabarui; all of them belong to tribe Chironomini, subfamily Chironominae, which was in accordance with traditional morphological classification.
Ethical approval
This study did not involve any ethical issues.
Author contributions
Yun-Li Xiao performed the analysis and wrote the paper. Jiaxin Wang performed the experiments, extracted DNA, and assembled the mitochondrial genome. Zi-Gang Xu annotated the mitochondrial genome. Xiang-Liang Fang carried out the phylogenetic analysis. Yue Fu drafted the manuscript, critically revised it for important content, and provided the final approval of the version for publication. The manuscript is approved by all authors for publication.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that was obtained at this study are available in the NCBI under the accession number MZ747090 (https://www.ncbi.nlm.nih.gov/nuccore/MZ747090). The associated BioProject, Bio-Sample numbers, and SRA are PRJNA804390, SAMN25889263, and SRR18003347, respectively.
Additional information
Funding
References
- Armitage PD, Cranston PS, Pinder LCV. 1995. The Chironomidae biology and ecology of non-biting midges. London: Chapman & Hall.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Cranston PS. 2007. The Chironomidae larvae associated with the tsunami-impacted waterbodies of the coastal plain of south-western Thailand. Bullet Raffles Museum. 55:231–244.
- Cranston PS, Ang YC, Heyzer A, Lim RBH, Wong WH, Woodford JM, Meier R. 2013. The nuisance midges (Diptera: Chironomidae) of Singapore’s Pandan and Bedok reservoirs. Raffles Bullet Zool. 61:779–793.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Jacobsen RE, Perry SA. 2007. Polypedilum nubifer, a chironomid midge (Diptera: Chironomidae) new to Florida that what has nuisance potential. Florida Entomologist. 90(1):264–267.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47(W1):W256–W259.
- Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Paine G, Gaufin AR. 1956. Aquatic Diptera as indicators of pollution in a Midwestern stream. Ohio J Sci. 56:291–304.
- Skuse FAA. 1889. Diptera of Australia. Part VI. The Chironomidae. Proc Linnean Soc New South Wales. 2:215–311.
- Tabaru Y, Moriya K, Ali A. 1987. Nuisance midges (Diptera: Chironomidae) and their control in Japan. J Am Mosq Control Assoc. 3(1):45–49.
- Trayler KM, Pinder AM, Davis JA. 1994. Evaluation of the juvenile hormone mimic pyriproxyfen (S-31183) against nuisance chironomids (Diptera: Chironomidae), with particular emphasis on Polypedilum nubifer (Skuse). Aust J Entomol. 33(2):127–130.
- Wang XH, Liu WB, Lin XL, Song C, Sun BJ, Yan CC, Qi X. 2020. Chironomidae. In: Yang D, Li Z, Liu QF, editors. Species catalogue of China, volume 2 animals, Insecta (V), Diptera (1), Nematocera. Beijing: Science Press.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.