Abstract
Viola kunawarensis Royle is a precious Uygur medicinal material that has anti-fever and detoxifying effects. This study reports the complete chloroplast genome sequence of V. kunawarensis based on Illumina NovaSeq-PE150 platform sequencing reads. The genome is 156,837 bp long and contains a small single-copy (SSC) region of 17,059 bp and a large single-copy (LSC) region of 86,194 bp, separated by two inverted repeats (IRs) of 26,792 bp each. There are 111 unique genes in the chloroplast genome. In this study, V. kunawarensis was confirmed to be most closely related to all comprising Viola taxa except Viola mirabilis and Viola websteri.
Viola kunawarensis Royle 1839 is a perennial herb widely distributed in the Himalayas, the Pamir region, Tianshan, and the Tibet and Xinjiang autonomous regions of China (Chinese Academy of Science Flora of China Editorial Board Citation1991). The whole plant of V. kunawarensis is officinal, and it is included in the processing specification of traditional Chinese medicine in Xinjiang Uygur Autonomous Region. In Uygur traditional medicine, V. kunawarensis was once named Viola tianshanica Maxim., and it has been used to treat fever, cold, pleurisy, pneumonia, pharyngitis, sore, swelling, and carbuncles, among other conditions (Yu Citation2020). Modern research shows that V. kunawarensis has medicinal effects that include anti-inflammatory and antibacterial (Liang et al. Citation2015), cough relief, anti-phlegm, anti-asthma (Qu et al. Citation2011), and anti-tumor effects (Cao et al. Citation2014; Ji and Wei Citation2015), which have attracted much attention in recent years.
At present, V. kunawarensis is the only source of medicinal materials which has been collected by “Processing specification of traditional Chinese medicine in Xinjiang Uygur Autonomous Region” in the genus Viola. Because of the limited availability and high price of the species, various species in Viola are used as substitutes for V. kunawarensis in the market, including Viola odorata and Viola philippica (Fan et al. Citation2019), which has seriously affected the safety and integrity of medicinal materials. However, owing to complex variation and interspecific heterogeneity, there are different opinions on the phylogeny of the genus Viola (Liang and Xing Citation2010; Sun et al. Citation2014). Therefore, it is extremely difficult to identify the basis of commercial V. kunawarensis medicinal materials by traditional morphological methods, which restricts the development of the industry. There were 11 species of published chloroplast genomes in the family Violaceae. In this study, the chloroplast genome of V. kunawarensis was assembled for the first time. The determination of this complete plastid genome sequence is a useful resource that will provide molecular data to illuminate its phylogenetic relationship within the genus and to better identify medicinal materials.
Plant materials of V. kunawarensis were collected from Taxkorgan County (75.4941°E, 37.2308°N), Xinjiang, China. The voucher specimen was deposited at the Xinjiang Institute of Chinese Materia Medica and Ethnical Materia (Urumqi, Congzhao Fan, [email protected]) with the voucher number 653131200614015LY. Genomic DNA was extracted using a DNeasy Plant Mini Kit (QIAGEN, Valencia, CA, USA) in this study. The Illumina NovaSeq-PE150 platform (Illumina, San Diego, CA, USA) was used to the genomic data of V. kunawarensis. The sequencing reads were assembled using MITObim v1.9 (Hahn et al. Citation2013), and the genome map was prepared using OGDraw online tools (Lohse et al. Citation2013).
The complete chloroplast genome was annotated using Viola raddeana (MH229818) as a reference. The chloroplast genome of V. kunawarensis has a circular quadripartite structure that resembles other angiosperm chloroplast genomes (Hahn et al. Citation2013). It has a length of 156,837 bp, containing a small single-copy (SSC) region of 17,059 bp and a large single-copy (LSC) region of 86,194 bp separated by two inverted repeats (IRs) of 26,792 bp each. The overall nucleotide composition is asymmetric (31.4% A, 32.4% T, 17.8% G, and 18.4% C) with AT content (63.8%) higher than GC content (36.2%).
The V. kunawarensis chloroplast genome contains 111 unique genes, including 77 protein-coding genes (PCGs), 4 rRNA genes, and 30 tRNA genes. In addition, 17 genes contain one or two introns, which includes 9 PCGs (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, and ycf3) possessing a single intron, 2 PCGs (clpP and ycf3) harboring two introns, and 6 tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) harboring a single intron.
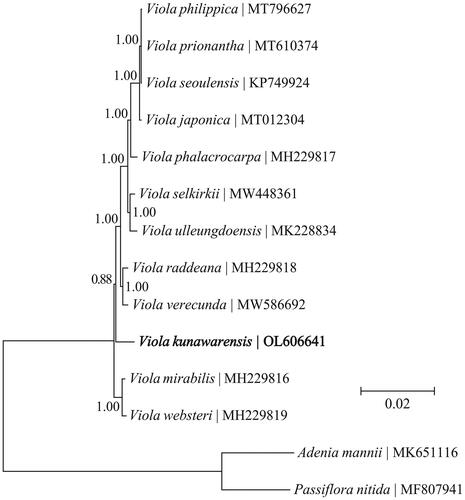
Geneious R11 was to. In total, 17 PCGs (atpA, atpB, matK, ndhA, ndhD, ndhF, ndhH, psaA, psaB, psbA, psbB, psbC, psbD, rbcL, rpoA, rpoB, and rpoC1) which yielded reliable alignments were selected for a panel of 14 taxa (incl. 12 ingroup and 2 outgroup taxa). These PCGs were individually aligned and were then concatenated into a single alignment in Geneious R11. After that, the resultant alignment was exported into TOPALi v2.5 (Milne et al. Citation2009), and a phylogenetic tree was constructed with the program MrBayes v3.1.2 (Ronquist and Huelsenbeck Citation2003) as implemented in TOPALi v2.5. It was revealed that V. kunawarensis was relatively basal in the genus Viola. It was basal to the clade comprising all Viola taxa except Viola mirabilis and Viola websteri, . These results would contribute to a better understanding of the genus Viola and provide molecular markers for the identification of V. kunawarensis and its congeners.
Ethical approval
In present study, V. kunawarensis plants were collected from the wild environment of Taxkorgan County, and the permission for collection of sample were obtained from Tashkurgan Wildlife Reserve Service Center. We complied with the Regulations of Xinjiang Uygur Autonomous Region on the protection of wild plants.
Author contributions
Congzhao Fan contributed to the conception of the study. Jizhao Zhang and Yuanjin Qiu performed the experiments and the data analysis. Jinchao Zhou performed the experiments and wrote the manuscript. All authors read and approved the final manuscript and agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
We confirm that the complete mitogenome sequence can be accessed via NCBI GenBank accession no. OL606641 at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA791966, SRR17326421, and SAMN24371149, respectively.
Figure 1. Phylogeny of the genus Viola based on the Bayesian phylogenetic analysis of the plastid protein-coding genes. The support values next to the nodes are Bayesian posterior probabilities according to the Bayesian analysis. The outgroup taxa used in this study are two species within the family Passifloraceae, i.e. Adenia mannii and Passiflora nitida.
Additional information
Funding
References
- Cao SW, Hu Y, Zhu XN, Chen RZ, Wang XL. 2014. The effect of esculetin on glioma growth in BALB/C nude: an in vivo study. J Sun Yat-Sen Univ Med Sci. 35(5):672–679.
- Chinese Academy of Science Flora of China Editorial Board. 1991. Flora of China. Beijing (China): Science Press; p. 8–29.
- Fan CZ, Zhu J, Zhao YQ, Wang GP, Li XJ. 2019. Study on the origin plant of Xinjiang ethnic medicines Viola kunawarensis based on DNA barcode technology. World Sci Technol Modern Tradit Chin Med. 21(3):553–558.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucl Acids Res. 41(13):e129–e129.
- Ji J, Wei ZM. 2015. Effects of aesculetin on the proliferation of human liver cancer cells BEL-7402 in vitro. Pharm Clin Chin Mater. 31(3):38–41.
- Liang CY, Zhang SY, Mao GN, Song HH, Ding SJ, Wang L, Chen XF. 2015. Advances in the synthesis and pharmacological activities of esculetin and its derivatives. J Shanxi Univ Sci Tech Nat Sci. 33(2):126–133.
- Liang GX, Xing FW. 2010. Infrageneric phylogeny of the genus Viola (Violaceae) based on trnL- trnF, psbA-trnH, rpL16, ITS sequences, cytological and morphological data. Acta Botanica Yunnanica. 32(6):477–488.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25(1):126–127.
- Qu AT, Sun C, Liu XY, Li LJ, Bai MAT, Zhang J. 2011. Mongolian heat pain shamisen soup powder aesculetin determination. Chin J Exp Tradit Med Form. 17(2):87–88.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Sun XQ, Liu M, Sun TH, Zhang XX, Shi CQ. 2014. Morphological study of the leaf structures of Viola in northeastern China and discussions of their taxonomic values (Violaceae). Acta Prataculturae Sinica. 23(2):223–234.
- Yu ZX. 2020. Processing specification of traditional Chinese medicine in Xinjiang Uygur Autonomous Region. Beijing (China): China Medical Science Press; p. 16.
