Abstract
Homalocladium platycladum is a fascinating ornamental plant that has long been used in Chinese medicine. Here, we characterize the complete chloroplast genome sequence of this plant (GenBank: NC_062330). This circular genome has a total length of 163,202 bp containing a large single-copy region (87,820bp), a small single-copy region (13,538bp), and a pair of inverted repeat regions (30,922bp). A total of 130 predicted genes were identified, including 85 protein-coding genes, 37 transfer RNA genes, and 8 ribosomal RNA genes. Phylogenetic analysis demonstrated that H. platycladum belongs to the Polygonaceae family and is highly analogous with Homalocladium and Muehlenbeckia families.
Homalocladium platycladum (F. Muell.) L.H. Bailey [syn. Muehlenbeckia platyclada (F. Muell.) Meisn.], as the only species of Homalocladium, Polygonaceae, originates in the Solomon Islands in the South Pacific (Jørgensen et al. Citation2015), F.J.H. von Mueller published Polygonum platycladum F. Muell. as a basionym in 1858 (Victoria Citation1858). In Southern China, H. platycladum is widely planted for its horticultural benefit as well as its therapeutic effects in the treatment of snake bites and fractures (Yen et al. Citation2009). Chloroplast genomes are often employed in various scientific pursuits, including plant phylogenetic analysis (Refulio-Rodriguez and Olmstead Citation2014) and plant identification (Nock et al. Citation2011). However, the chloroplast genome of H. platycladum has not been formally analyzed and reported in the literature. Here, for the first time, we report the complete chloroplast genome sequence of H. platycladum and its phylogenetic analysis.
Samples of H. platycladum used for this study were collected at the Institute of Botany, Chinese Academy of Sciences (IBCAS), Beijing(116° 21′ N, 39° 99′ E), and the specimen was identified by Wei Zhai. A specimen was deposited at the Herbarium of Anqing Normal University (contact person: Wei Zhai, email: [email protected]) under the voucher number ZW2901. The experimental material is not an endangered species and is cultivated by individuals, and no approval is required for the collection of specimens. The chloroplast genome DNA library was constructed and sequenced on the Illumina NovaSeq 6000 (Novogene Co. Ltd, Beijing, China) according to the manufacturer’s instructions. Run reports detailed a total of ∼11.5 GB of clean reads which were then assembled using SPAdes v3.15.3 (Bankevich et al. Citation2012) and Bandage (Wick et al. Citation2015). Total coverage of the sequencing was evaluated using BWA (Li and Durbin Citation2009) after initial de novo assembly of the chloroplast genome sequence, and the mean genome coverage: was 1435.2×. Assembled chloroplast genome sequences were annotated using PGA (Qu et al. Citation2019). Finally, annotated results were manually corrected using Geneious R11.1 software (Kearse et al. Citation2012) to ensure correctness. The annotated chloroplast genome sequence of H. platycladum has been submitted to GenBank and received the accession number NC_062330.
The total length of the H. platycladum chloroplast genome is 163,202 bp, containing a large single-copy (LSC) region of 87,820 bp, a small single-copy (SSC) region of 13,538 bp, and a pair of inverted repeat regions (IRa and IRb) of 30,922 bp each. The overall GC content of the genome was 37.3%. A total of 130 predicted genes were identified, including 85 protein-coding genes (CDS), 37 transfer RNA genes (tRNA), and 8 ribosomal RNA (rRNA) genes. Among the annotated genes, 6 protein-coding, 7 tRNA, and 4 rRNA genes were duplicated in the inverted repeat regions. Fifteen genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained only a single intron, while four genes (ycf3, clpP, and two rps12) possess two introns.
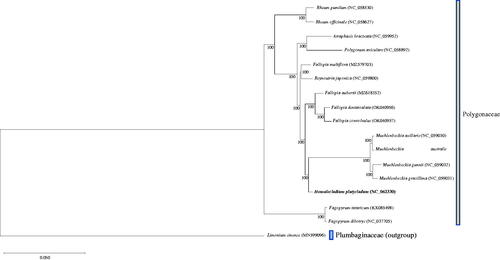
Fifteen available complete chloroplast genomes from Polygonaceae were selected based on annotation quality and sequence homology to indicate the phylogenetic position of H. platycladum and Limonium sinense (MN599096) from Plumbaginaceae, the sister family of Polygonaceae (Group et al. Citation2016), was selected as an outgroup. Chloroplast sequences from the different species were aligned by MAFFT v7.490 (Katoh and Standley Citation2013), and the maximum-likelihood (ML) phylogenetic tree was constructed in IQ-TREE v1.6.12 (Nguyen et al. Citation2015), with 1000 bootstrap replicates. The phylogenetic tree indicates that H. platycladum belongs to the Polygonaceae family and supports a sister relationship between Homalocladium and Muehlenbeckia with high bootstrap value (). In total, this study illustrates the chloroplast genome structure of H. platycladum and gives crucial information for understanding its evolutionary development.
Ethical approval
We did not need permission to use these samples because this research does not include ethical research and all experimental materials were personally grown by Wei Zhai.
Author contributions
Zijie Chen: assembly and analysis of chloroplast genome data and Original draft preparation; Wei Zhai and Kai Zhao: formulation of overarching research goals and preparation of experimental materials and apparatus; Qingxiang Fu and Changrui Tai: critical review, commentary and revision; All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data of H. platycladum that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. NC_062330. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA824631, SRS12542718, and SAMN27409548, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Byng JW, Chase MW, Christenhusz MJM, Fay MF, Judd WS, Mabberley DJ, Sennikov AN, Soltis DE, Soltis PS, Stevens PE, et al. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181(1):1–20.
- Jørgensen PM, Nee MH, Beck SG. 2014. Catálogo de las plantas vasculares de Bolivia. Missouri Botanical Garden Press. 127(1–2): i–viii, 1–1744.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li H, Durbin RJB 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25(14):1754–1760.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Nock CJ, Waters DLE, Edwards MA, Bowen SG, Rice N, Cordeiro GM, Henry RJ. 2011. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol J. 9(3):328–333.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Refulio-Rodriguez NF, Olmstead RG. 2014. Phylogeny of lamiidae. Am J Bot. 101(2):287–299.
- Mueller F. 1858. Transactions of the Philosophical Institute of Victoria from January to December. Philosophical Institute of Victoria. (Vol. 2):73.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Yen C-T, Hsieh P-W, Hwang T-L, Lan Y-H, Chang F-R, Wu Y-C. 2009. Flavonol glycosides from Muehlenbeckia platyclada and their anti-inflammatory activity. Chem Pharm Bull. 57(3):280–282.