Abstract
To better understand the taxonomy of the genus Oxytropis, we sequenced the complete chloroplast genome of Oxytropis aciphylla Ledeb. The total plastome of O. aciphylla Ledeb. is 122,121 bp in length with a GC content of 34.3%. It contains one large single-copy (LSC) region of 88,235 bp, one small single-copy (SSC) region of 10,400 bp, and one inverted repeat (IR) region of 23,486 bp, encoding 76 proteins, four rRNAs, and 29 tRNAs. The phylogenetic position shows that O. aciphylla Ledeb. is the closest to Oxytropis glabra.
Oxytropis is one of the largest genera of the Leguminosae family (Malyshev Citation2008). It includes approximately 450 species and distributed in Inner Mongolia, Shaanxi, Ningxia, Gansu, Qinghai, Xinjiang, and other provinces of China, as well as in Western Siberia of Russia and southern Mongolia. It has been reported that Central Asia and West Asia are the most important centers for the speciation of Oxytropis (Kholina et al. Citation2021). However, due to the high diversity of Oxytropis, the taxonomy of this genus is poorly understood. The complete chloroplast genome barcode not only plays an important role in the identification of plant species, but also plays a crucial role in the taxonomic relationship of poorly studied species (Techen et al. Citation2014; Tekpinar et al. Citation2016). However, the current research on the entire chloroplast genome of Oxytropis species is still poor.
Oxytropis aciphylla Ledeb. is a dwarf cushion-shaped leguminous perennial subshrub in the genus Oxytropis (Ledebour Citation1831). It mainly grows on hill slopes and sandy Gobi deserts (Zheng et al. Citation2013). As a desert plant, it is popular for cattle and sheep in the early spring and is the major plant litter of sandy Gobi deserts (Li et al. Citation2015). The whole chloroplast genome information of O. aciphylla Ledeb. has not been reported in the NCBI database. Therefore, in this study, we sequenced and structurally characterized its chloroplast genes, aiming to provide a useful resource for future research on Oxytropis genetic evolution.
Fresh leaves of O. aciphylla Ledeb. were collected from the Gobi (38°08′53.92″ N,105°54′29.08″ E) of Helan Mountain in Yinchuan City, Ningxia Hui Autonomous Region. Total genomic DNAs were extracted from the fresh leaves using the CTAB method (Doyle and Doyle Citation1987). A library with an average length of 150 bp was constructed using the Nextera XT DNA library preparation reagents (Illumina, San Diego, CA) and sequenced by the Illumina NovaSeq 6000 platform. A total of 6.01 Gb of sequence reads were generated and edited using NGS QC Toolkit v2.3.3. Contigs were obtained from high-quality reads using the de novo assembler SPAdes 3.11.0 software (Bankevich et al. Citation2012) and annotated using plan software (Huang and Cronk Citation2015) with Oxytropis glabra as the assembly and annotation reference genome. The complete sequence was submitted to GenBank (OK143433), and the sample was stored at the Laboratory of Ecosystem, North Minzu University, Yinchuan (voucher specimen: NMU00047) (Zhanglei, email: [email protected]).
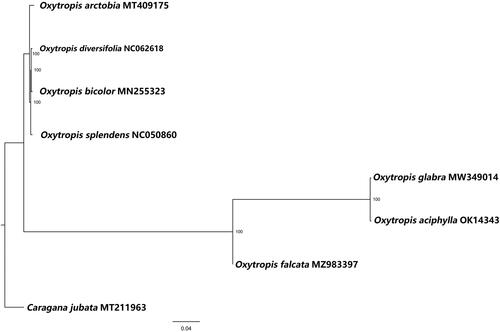
The complete chloroplast genome of O. aciphylla Ledeb. is 122,121 bp in length with an overall GC content of 34.3%. It contains an 88,235 bp large single-copy (LSC) region, a 10,400 bp small single-copy (SSC) region, and a 23,486 bp inverted repeat (IR) region and encodes 109 genes, including 76 PCGs, four rRNA, and 29 tRNA. To confirm the phylogenetic position of O. aciphylla Ledeb. and understand its relationship with other species in Leguminosae, the complete chloroplast genomes of seven species in the Leguminosae family, including Oxytropis glabra (NC056975.1) (Liu et al. Citation2021), were collected and aligned with O. aciphylla Ledeb. Subsequently, a phylogenetic tree was constructed by IQTREE v1.6 with 1000 bootstraps replicates using the Best-fit model (Nguyen et al. Citation2015; Hoang et al. Citation2018). A final ML tree was constructed using Caragana jubata as the outgroup. As shown in , the entire tree was separated into two major clades: Oxytropis, and Caragana, with one Caragana species branch, seven Oxytropis species branches. O. aciphylla Ledeb. has the closest relationship with Oxytropis glabra. We believe that our results will be highly beneficial to studies on species identification, phylogenetic relationships, and population genetics of Oxytropis species as well as evolution in the Leguminous family.
Figure 1. Maximum-likelihood phylogenetic tree for Oxytropis aciphylla Ledeb. based on complete chloroplast genomes of seven species in family Leguminosae, with Caragana jubata as the outgroup. The numbers to the right of the branches are bootstrap support values. At the bottom of the figure is the distance scale of phylogenetic tree.
Ethics statement
The samples were collected in compliance with the Regulations of the People’s Republic of China on Wild Plants Protection promulgated by Decree No. 204 of the State Council of the People’s Republic of China on September 30, 1996.
Author contributions
Zhanlin Bei: data curation, formal analysis, and writing-original draft; Lei Zhang: resources and supervision; Xingjun Tian: conceptualization, funding acquisition, resources, supervision, writing-review and editing.
Disclosure statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability statement
The chloroplast genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number OK143433. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA769668, SRR16249213, and SAMN22161435, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Kholina A, Kozyrenko M, Artyukova E, Sandanov D, Selyutina I. 2021. Genetic diversity of Oxytropis section Xerobia (Fabaceae) in one of the centres of speciation. Genetica. 149(2):89–101.
- Ledebour KF. 1831. Oxytropis aciphylla. Flora Altaica. 3:279–280.
- Li X, Liu B, Chen L, Song N. 2015. Effects of litter accumulation on plant communities in fenced desert steppe. Pol J Ecol. 63(3):333–340.
- Liu S, Wei Y-RL, Si W, Qu W-R, Yang T-G, Wu Z-H, Jiao P-P. 2021. Complete chloroplast genome sequence of Oxytropis glabra (Leguminosae). Mitochondrial DNA B Resour. 6(9):2478–2479.
- Malyshev LI. 2008. Phenetics of the subgenera and sections in the genus Oxytropis DC. (Fabaceae) bearing on ecology and phylogeny. Contemp Probl Ecol. 1(4):440–444.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Techen N, Parveen I, Pan Z, Khan IA. 2014. DNA barcoding of medicinal plant material for identification. Curr Opin Biotechnol. 25:103–110.
- Tekpinar A, Karaman Erkul S, Aytaç Z, Kaya Z. 2016. Phylogenetic relationships among native Oxytropis species in Turkey using the trnL intron, trnL-F IGS, and trnV intron cpDNA regions. Turk J Bot. 40(5):472–479.
- Zheng JG, Chen YW, Wu GX. 2013. Association of vegetation patterns and environmental factors on the arid western slopes of the Helan Mountains, China. Mt Res Dev. 33(3):323–331.
