Abstract
Zanthoxylum esquirolii Léveillé Citation1914 is mainly distributed in southwest China, and its wild germplasm resources are scarce and in urgent need of conservation. In this study, we report the first complete chloroplast genome sequence of Z. esquirolii using next-generation sequencing. The circular genome is 158,390 bp in length, containing two inverted repeat (IR) regions of 27,622 bp separated by a large single copy (LSC) region of 85,580 bp and a small single copy (SSC) region of 17,566 bp. The chloroplast genome contains a total of 132 genes, including 87 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall GC content of the chloroplast genome was 38.46%, with corresponding values in the LSC, SSC, and IR regions of 36.84%, 33.55%, and 42.51%, respectively. The phylogenetic tree revealed that Z. esquirolii Levl. formed a clade with Z. piperitum DC., Z. bungeanum Maxim., Z. simulans Hance and Z. sp. NH-2018, and had a strongly supported sister relationship with Z. bungeanum.
In 1914, Léveillé H. first described of Zanthoxylum esquirolii Levl. (Léveillé Citation1914). Z. esquirolii is an important economic forest tree species with good development and utilization value and is an excellent medicinal tree species mainly distributed in the provinces of Guizhou, Sichuan, Tibet and Yunnan in China (Huang, Citation1997). Z. esquirolii has the functions of warming the middle and dispelling cold, promoting blood circulation and relieving pain, and treating cold-related aches and pains, bruises, blood stasis, swelling and pain (He et al., Citation2011). This plant has great potential for use as a feedstock and medicine. However, the natural habitat of the species’s fragmented, and wild resources of Z. esquirolii have been dramatically depleted and need urgent conservation. A large body of knowledge regarding its genetic information would contribute to the formulation of a protection strategy. No genomic information of Z. esquirolii has been reported to date. In this study, we present the first complete chloroplast genome sequence of Z. esquirolii and construct its phylogenetic relationships with related species based on Illumina paired-end sequencing data. These data will provide a reference for the development and protection of germplasm resources in the future.
The fresh leaves of a single individual of Z. esquirolii were collected from Xishui County, Guizhou, China (28.6712° N, 106.4561° E), and a voucher specimen was deposited at the Chongqing University of Arts and Sciences Herbarium (GZLX1) under accession number CUAS-GZ20180720 (Xia Liu, [email protected]). Genomic DNA was extracted using a modified CTAB method (Doyle Citation1987). The DNA library was sequenced by Hefei Bio&Data Biotechnologies Inc. (Hefei, China) on the BGISEQ-500 platform with PE150 read lengths. The clean reads were used for the de novo assembly of the chloroplast genome using SPAdes Assembler v3.9.0 (Bankevich et al. Citation2012). With Z. bungeanum Maxim. (NC_031386) as the reference. The annotation of the complete genome was performed using CpGAVAS (Liu et al. Citation2012) and GeSeq software (Michael et al. Citation2017). After a manual check and adjustment, the annotated chloroplast genome sequence of Z. esquirolii was submitted to GenBank (MZ676709).
The chloroplast genome of Z. esquirolii is a double stranded, circular DNA 158,390 bp in length that contains two inverted repeat (IR) regions of 27,622 bp separated by a large single-copy (LSC) region and a small single-copy (SSC) region of 85,580 bp and 17,566 bp, respectively. The chloroplast genome encodes a total of 132 genes (87 protein-coding, 37 tRNA, and 8 rRNA genes), with 18 duplicated genes (7 protein-coding, 7 tRNA, and 4 rRNA genes). Nineteen genes contain two exons and four protein-coding genes (ycf3, clpP, and two rps12) contain three exons. The overall GC content of Z. esquirolii is 38.46% and the values in the LSC, SSC and IR regions are 36.84%, 33.55%, and 42.51%, respectively.
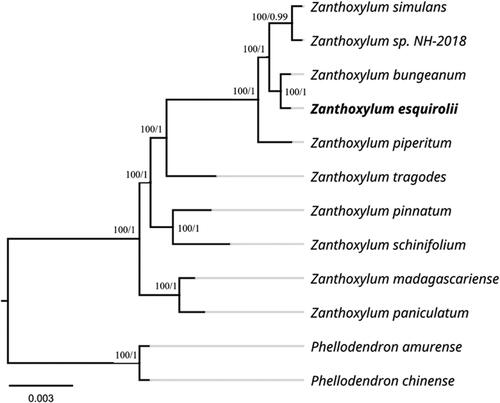
The phylogenetic analysis was performed using 12 complete plastid genomes, with Phellodendron amurense Rupr. and Phellodendron chinense Schneid. as the outgroup (). The 12 complete chloroplast genome sequences were subjected to multiple sequence alignment using MAFFT software (Katoh and Standley Citation2013). The best models of the complete chloroplast genomes for the ML and BI phylogenetic analyses were determined by jModelTest 2.1.1 (Posada Citation2008) with an Akaike Information Criterion (AICc). The best fitting evolutionary model for the combined complete chloroplast genome dataset was TVM + I + G in the ML and Bayesian analyses. A maximum likelihood (ML) phylogenetic tree was built using the RAxML version 8 program (Alexandros Citation2014) with 1000 bootstrap replicates. For BI in MrBayes 3.1.2 (Ronquist and Huelsenbeck Citation2003), two independent Markov chain Monte Carlo (MCMC) runs were performed and contained four MCMC chains that were run for 1000,000 generations and sampled every 1000 generations; all other parameters were set to default. The first 25% of the sampled trees were abandoned as burn-in to check the stability of each run, and the posterior probabilities (PP) were calculated from the remaining trees. All phylogenetic trees were viewed using the Figtree v1.4.2 program (Rambaut Citation2014). The phylogenetic trees generated by the ML and Bayesian methods were most similar to each other but with different branch support values in some clades (). Phylogenetic analysis showed that the Zanthoxylum species formed a monophyletic group. Z. esquirolii is most closely related to Z. bungeanum and is sister to Z. sp. NH-2018 and Z. simulans Hance, with 100% bootstrap support.
Figure 1. Maximum-likelihood phylogenetic tree of Z. esquirolii and other related species based on complete chloroplast genome sequences. Numbers near the nodes sequentially indicate ML/BI support values. The following sequences were used: Zanthoxylum simulans Hance NC_037482 (Hou et al. Citation2017), Zanthoxylum sp. NH-2018 MF716521, Zanthoxylum bungeanum Maxim NC_031386 (Liu and Wei Citation2017), Zanthoxylum esquirolii Levl. MZ676709, Zanthoxylum piperitum Maxim NC_027939 (Lee et al. Citation2016), Zanthoxylum tragodes NC_046747, Zanthoxylum pinnatum NC_046746, Zanthoxylum schinifolium Sieb. et Zucc. NC_046746, Zanthoxylum madagascariense NC_046744, Zanthoxylum paniculatum NC_046745, Phellodendron amurense Rupr. NC_035551 and Phellodendron chinense Schneid. MT916287.
The limited number of polymorphic loci produced by low-resolution markers hinders the phylogenetic research on Zanthoxylum species in previous studies (Medhi et al. Citation2014; Feng et al. Citation2015; Kim et al. Citation2017; Appelhans et al. Citation2018). The emergence of the chloroplast genomes of a large number of Zanthoxylum species can provide important insights into the evolution of Zanthoxylum in eastern Asia. This complete chloroplast genome can be used for phylogenetic, population, and chloroplast genetic engineering studies of Z. esquirolii and is fundamental for the creation of new conservation and management strategies for this important medicinal plant species.
Ethical approval
The present study was approved by the authors’ institution (the Chongqing University of Arts and Sciences) and national. The research does not involve a threatened/endangered species. All the research meets ethical guidelines and adheres to the legal requirements of the study country. The collection of plant material has been carried out in accordance with the International Union for Conservation of Nature (IUCN) policies research involving species at risk of extinction, the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Author contributions
Xia Liu, Qinqin Huang and Zexiong Chen designed the study, writing and revised the manuscript; Chong Sun and Han Liu involved in the process of sequences editing and phylogenetic analyses; Can He, Fengting Huang and Haowen Liu participated in the collection and identification of plant material. All authors read and approved the final manuscript, and agreed to be accountable for all aspects of the work.
Acknowledgements
We thank Xia Gong, Yinming Wu and Zheng Chen (Sichuan Academy of Botanical Engineering, China) for their help in sample collections. We kindly thank the anonymous reviewers for their helpful comments and corrections, which have helped us to improve the submitted manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ676709. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA680256, SRR17163937, and SAMN23766595, respectively.
Additional information
Funding
References
- Alexandros S. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Appelhans MS, Reichelt N, Groppo M, Paetzold C, Wen J. 2018. Phylogeny and biogeography of the pantropical genus Zanthoxylum and its closest relatives in the proto-Rutaceae group (Rutaceae). Mol Phylogenet Evol. 126:31–44.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Huang CJ. 1997. Rutaceae. Flora Reipublicae Popularis Sincecae. Vol. 43. Beijing: Science Press.
- Feng S, Yang T, Li X, Chen L, Liu Z, Wei A. 2015. Genetic relationships of Chinese prickly ash as revealed by ISSR markers. Biologia. 70(1):45–51.
- He SZ, Tang XG, Xu WF, Liu MT. 2011. Study on species and geographic distribution of the medicinal plants resources of Zanthoxylum in Guizhou. Res Pract Chin Med. 25(4):27–30.
- Hou N, Wang G, Feng SJ, Wei AZ. 2017. The complete chloroplast genome of an aromatic Chinese pepper (Zanthoxylum simulans). Mitochondrial DNA B Resour. 3(1):26–27.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim YM, Jo A, Jeong JH, Kwon YR, Kim HB. 2017. Development and characterization of microsatellite primers for Zanthoxylum schinifolium (Rutaceae). Appl Plant Sci. 5(7):1600145.
- Léveillé H. 1914. Zanthoxylum esquirolii. Repert Spec Nov Regni Veg. 13(363–367):266.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Liu YL, Wei AZ. 2017. The complete chloroplast genome sequence of an economically important plant, Zanthoxylum bungeanum (Rutaceae). Conservation Genet Resour. 9(1):25–27.
- Lee J, Lee HJ, Kim K, Lee SC, Sung SH, Yang TJ. 2016. The complete chloroplast genome sequence of Zanthoxylum piperitum. Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3525–3526.
- Michael T, Pascal L, Tommaso P, Elena SUJ, Axel F, Ralph B, Stephan G. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Medhi K, Sarmah DK, Deka M, Bhau BS. 2014. High gene flow and genetic diversity in three economically important Zanthoxylum Spp. of upper Brahmaputra valley zone of NE India using molecular markers. Meta Gene. 2:706–721.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256.
- Rambaut A. 2014. FigTree v1.4.2. Edinburgh: University of Edinburgh.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
