Abstract
We have sequenced the Ziziphus jujuba cv. Bokjo chloroplast genome by de novo assembly using next-generation sequencing. The complete circular chloroplast genome consisted of 161,714 bp and contained four parts: a large single-copy (LSC) region of 89,323 bp, a small single-copy (SSC) region of 19,361 bp, and two inverted repeat regions (IRa and IRb) of 26,515 bp each. The genome annotation predicted a total of 110 genes, including 76 protein-coding genes, 30 tRNA genes, and four rRNA genes. Phylogenetic analysis demonstrated the close taxonomic relationship between Z. jujuba cv. Bokjo and two other members of the Ziziphus genus, Z. spina-christi and Z. mauritiana. We found 135 polymorphic loci, 63 single nucleotide polymorphism (SNP) and 72 insertion–deletion (InDel), from the comparison of Z. jujuba cultivar Bokjo and Z. jujuba reference (NC_030299). The polymorphic loci could be used for the differentiation of Z. jujuba genetic resources and for breeding in the future.
Jujube (Ziziphus jujuba Mill. 1768) is a deciduous tree belonging to the genus Ziziphus of the Rhamnaceae family. Z. jujuba inhabits subtropical and tropical arid regions, but some species occur in temperate and arid regions with continental climates (Evreinoff Citation1964; Nam et al. Citation2015). Z. jujuba extract contains the alkaloids lysicamine and nornuciferine and is an effective sedative (Han and Park Citation1987). A long period of evolution and artificial selection has given rise to various varieties and cultivars of jujube, and more than 800 cultivars have been reported (Wang et al. Citation2014). Despite the economic importance of jujube, molecular research on the species is limited, as is knowledge concerning its genetic diversity and cultivar identification (Liang et al. Citation2019).
We have completed the chloroplast genome of Z. jujuba cv. Bokjo, the most widely cultivated jujube cultivar in Korea (Lee et al. Citation2018). Z. jujuba cv. Bokjo leaf samples for DNA extraction were collected from Chungcheongbuk-do Agricultural Research and Extension Service (CBARES) (36°34′38.7″N, 127°44′52.9″E). A dried plant specimen was deposited in the Herbarium of the National Institute of Horticulture and Herbal Science, Eumsung, Republic of Korea (http://www.nihhs.go.kr/; Contact: Yoongee Lee; [email protected]) under the voucher number MPS006272. Total genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA). The isolated genomic DNA was used to construct a paired-end (PE) library with a mean insert size of 700 bp, using the Illumina HiSeq platform. Contigs were assembled with a total of 11.96 Gbp PE reads collected from Illumina Hiseq next-generation sequencing using the CLC Genomics Workbench (ver. 11.0, Qiagen, Aarhus, Denmark). The assembled structure and the genes of the complete chloroplast genome were annotated using the DOGMA program (http://dogma.ccbb.utexas.edu/) (Wyman et al. Citation2004) and were manually corrected based on a BLAST search. The Z. jujuba cv. Bokjo complete chloroplast genome sequence was deposited in GenBank under accession no. MT919946. The complete Z. jujuba cv. Bokjo chloroplast genome was a circular molecule with a length of 161,714 bp and a GC content of 36.75%, and was composed of four distinct regions: a large single-copy region of 89,323 bp, a small single-copy region of 19,361 bp, and two inverted repeat regions (IRa and IRb) each of 26,515 bp. We annotated 110 genes within the Z. jujuba cv. Bokjo chloroplast genome, which included 76 protein-coding genes, 30 tRNA genes, and four rRNA genes.
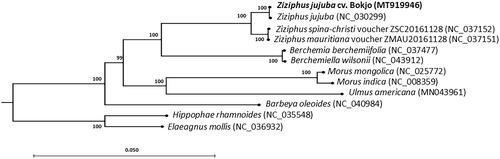
We performed a phylogenetic analysis based on the complete chloroplast genome sequence of Z. jujuba cv. Bokjo and of 11 other species, including five ingroup species of the Rhamnaceae to which Ziziphus belongs, and six outgroup species of the Rosales. To generate the phylogenetic tree, we used the time-reversal model of the maximum-likelihood algorithm with 1000 bootstrap replications in the CLC Genomics Workbench (ver. 11.0, CLC Qiagen). Six taxa of Rhamnaceae formed a monophyletic group that was distinct from six other species of the Rosales (). The phylogenetic tree placed Z. jujuba cv. Bokjo together with Z. jujuba (NC_030299) and close to Z. spina-christi and Z. mauritiana.
Figure 1. Maximum-likelihood phylogenetic tree based on the chloroplast genome sequence from Z. jujuba cv. Bokjo and related species in the Rhamnaceae family, to which the genus Ziziphus belongs. The chloroplast sequences of members of the Elaeagnaceae, Barbeyaceae, Moraceae, and Ulmaceae families were used as the outgroup. Numbers at each node represent the bootstrap values for 1000 replicates.
We compared the chloroplast genome of Z. jujuba cultivar Bokjo to a Z. jujuba reference (NC_030299) and we found 135 polymorphic loci, 63 single nucleotide polymorphisms (SNPs) and 72 InDels. The polymorphic loci could be used for the differentiation of Z. jujuba genetic resources and for breeding in the future.
Ethical approval
Ziziphus jujuba cultivar Bokjo were collected from the Chungcheongbuk-do Agricultural Research and Extension Service (CBARES) in Boeun, Republic of Korea, with permission from the CBARES researcher, Ms. Ha Kyung Oh. The plant is freely accessible to Ha Kyung Oh with non-commercial resource purpose. The authors comply with relevant institutional, national, and international guidelines and legislation for plant study.
Author contributions
H.K.O., K.H.L., J.L., and Y.L. conceived and designed the study. J.H.P., J.S.K., H.T.K., M.L., and J.L. analyzed the data. M.K., J.G., and M.L. performed the experiments. M.K., J.L., and Y.L. wrote the manuscript. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MT919946. The associated ‘BioProject’, ‘SRA’, and ‘Bio-Sample’ numbers are PRJNA794271, SRR17438409, and SAMN24619238, respectively.
Additional information
Funding
References
- Evreinoff VA. 1964. Notes sur le jujubier (Zizyphus sativa G.). J d'agric trop bot appl. 11(5):177–187.
- Han BH, Park MH. 1987. Sedative activity and its active components of Zizyphi fructus. Arch Pharm Res. 10(4):208–211.
- Lee J-E, Yun JH, Lee AR, Kim SS. 2018. Volatile components and sensory properties of jujube wine as affected by material preprocessing. Int J Food Prop. 21(1):2052–2061.
- Liang T, Sun W, Ren H, Ahmad I, Vu N, Huang J. 2019. Genetic diversity of Ziziphus mauritiana germplasm based on SSR markers and ploidy level estimation. Planta. 249(6):1875–1887.
- Nam JI, Kwon HY, Kim MS, Kim SH. 2015. Flowering characteristics and honeybee visiting of jujube (Ziziphus jujuba Mill). Apiculture. 30(4):343–348.
- Wang S, Liu Y, Ma L, Liu H, Tang Y, Wu L, Wang Z, Li Y, Wu R, Pang X. 2014. Isolation and characterization of microsatellite markers and analysis of genetic diversity in Chinese jujube (Ziziphus jujuba Mill.). PLOS One. 9(6):e99842.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
