Abstract
This study describes the complete mitochondrial genome sequence of the scyphozoan Acromitus flagellatus (Maas, 1903), a blooming jellyfish found in the coastal areas of Hainan, China. Its mitochondrial DNA is 16,779 bp in length and has a linear structure, comprising 13 protein-coding genes (PCGs), two rRNAs (s-rRNA and l-rRNA), and two tRNAs (trna-W-TCA and trna-M-CAT). A + T content was 65.39% (A: 29.27%, C: 16.59%, G: 18.03%, and T: 36.12%). ATG was the start codon in 11 PCGs: COX1, COX2, ATP8, ATP6, COX3, NAD2, NAD6, NAD4l, NAD1, NAD4, and COB. NAD5 and NAD3 had GTG as the start codon. TAG was the stop codon for COX2, NAD6, and COB. The other 10 PCGs were terminated by TAA. The neighbor-joining phylogenetic tree of the 15 related jellyfish species showed that A. flagellatus is closely related to Nemopilema nomurai and Rhopilema esculentum.
In recent decades, jellyfish blooms have frequently appeared in coastal seas worldwide (Condon et al. Citation2013). Jellyfish blooms cause serious damage to coastal economic development and marine ecosystem, and pose a challenge to human safety (Purcell Citation2007; Richardson et al. Citation2009). Extensive studies have examined the mechanisms of jellyfish blooms by focusing on understanding the eco-physiological characteristics of their life cycle stages (Lucas et al. Citation2012; Purcell Citation2012; Pitt et al. Citation2018). However, few studies have focused on the molecular phylogenetic relationships among jellyfish species based on their mitochondrial genomes (Zou et al. Citation2012; Hwang et al. Citation2014; Feng et al. Citation2019). This is important as it may provide insight into the similarity between different species of jellyfish.
The scyphozoan Acromitus flagellatus (Maas, 1903) belongs to Catostylidae (Family) under the Suborder Dactyliophorae. In recent years, medusae of A. flagellatus appeared in large numbers in the coastal waters of Hainan Province, China, from April to June (Du et al. Citation2021). To determine the molecular phylogenetic relationships between A. flagellatus and other jellyfish, its complete mitochondrial DNA was sequenced. An A. flagellatus medusa was collected from the eastern coast of Hainan Province (19.57° N, 110.83° E) and frozen in an ice box containing dry ice. The sample was then transferred to a −80 °C freezer at the Institute of Oceanology, Chinese Academy of Sciences (http://english.qdio.cas.cn/, Song Feng: [email protected]) under the voucher ‘A. flagellatus ①.’ A piece of muscle tissue from A. flagellatus medusa was used for extraction of mitochondrial DNA. The extracted DNA was also stored in a −80 °C freezer, as described above, for sequencing (voucher: DNA-A. flagellatus ①).
The complete mitochondrial genome of A. flagellatus showed a linear molecular structure with a length of 16,779 bp (GenBank accession no. OM457248.1). There were 13 protein-coding genes (PCGs), small and large subunit ribosomal RNAs (s-rRNA and l-rRNA), and methionine and tryptophan transfer RNAs (trna-W-TCA and trna-M-CAT) in the mitochondrial genome. The base composition was 29.27%, 18.03%, 16.59%, and 36.12% for A, G, C, and T, respectively. A + T base composition was less than 70% (65.39%), similar to that in Nemopilema nomurai and Rhopilema esculentum (Wang and Sun Citation2017a, Citation2017b).
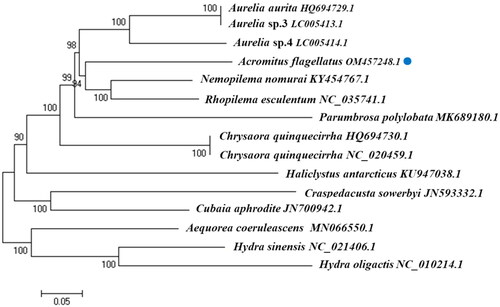
Of the 13 PCGs in A. flagellatus mitochondrial genome, 11 started with ATG (COX1, COX2, ATP8, ATP6, COX3, NAD2, NAD6, NAD4l, NAD1, NAD4, and COB). Only NAD5 and NAD3 had GTG as the start codon. Complete stop codons were present in all the genes. TAG was the stop codon for COX2, NAD6, and COB. The other 10 PCGs terminated with TAA as the stop codon. The neighbor-joining phylogenetic tree of 15 jellyfish species constructed using the complete mitochondrial genome from NCBI (), showed that A. flagellatus was closely related to N. nomurai (GenBank no. KY454767.1) and R. esculentum (GenBank no. NC_035741.1).
Ethical approval
The experiments were performed in accordance with the recommendations of the Ethics Committee of the Institute of Oceanology, Chinese Academy of Sciences. These policies were enacted in accordance with the Guide for the Care and Use of Laboratory Animals of the Chinese Association for Laboratory Animal Sciences (No. 2011-2). Permission was granted by the National Natural Science Foundation of China, Natural Science Foundation of Shandong Province, and Fundamental Research Funds of Shandong University.
Author contributions
SF and LW designed the study. QY collected the jellyfish samples and JL conducted the experiments. SF and LW analyzed the data. JL wrote the manuscript with the help of SF and LW.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov/) accession no. OM457248.1. The associated BioProject, SRA, and Bio-Sample numbers were PRJNA817707, SRR18358023, and SAMN26806304, respectively.
Additional information
Funding
References
- Condon RH, Duarte CM, Pitt KA, Robinson KL, Lucas CH, Sutherland KR, Mianzan HW, Bogeberg M, Purcell JE, Decker MB, et al. 2013. Recurrent jellyfish blooms are a consequence of global oscillations. Proc Natl Acad Sci USA. 110(3):1000–1005.
- Du C, He J, Sun T, Wang L, Wang F, Dong Z. 2021. Molecular identification on the causative species jellyfish blooms in the northern South China Sea in 2019. J Trop Oceanogr. 41:142–148.
- Feng S, Lin J, Sun S, Liu Q. 2019. Complete mitochondrial genome sequence of Parumbrosa polylobate. Mitochondrial DNA Part B. 4(2):2262–2263.
- Hwang DS, Park E, Won YJ, Lee JS. 2014. Complete mitochondrial genome of the moon jellyfish, Aurelia sp. nov. (Cnidaria, Scyphozoa). Mitochondrial DNA. 25(1):27–28.
- Lucas CH, Graham WM, Widmer C. 2012. Jellyfish life histories: role of polyps in forming and maintaining scyphomedusa populations. Adv Mar Biol. 63:133–196.
- Pitt K, Lucas CH, Condon RH, Duarte CM, Stewart-Koster B. 2018. Claims that anthropogenic stressors facilitate jellyfish blooms have been amplified beyond the available evidence: a systematic review. Front Mar Sci. 5:451.
- Purcell JE. 2012. Jellyfish and ctenophore blooms coincide with human proliferations and environmental perturbations. Ann Rev Mar Sci. 4:209–235.
- Purcell JE, Uye S, Lo W. 2007. Anthropogenic causes of jellyfish blooms and their direct consequences for humans: a review. Mar Ecol Prog Ser. 350:153–174.
- Richardson AJ, Bakun A, Hays GC, Gibbons MJ. 2009. The jellyfish joyride: causes, consequences and management responses to a more gelatinous future. Trends Ecol Evol. 24(6):312–322.
- Wang Y, Sun S. 2017a. Complete mitochondrial genome of the jellyfish, Nemopilema nomurai (Cnidaria: Scyphozoa) and the phylogenetic relationship in the related species. Mitochondrial DNA Part B. 2(1):165–166.
- Wang Y, Sun S. 2017b. Complete mitochondrial genome of the jellyfish, Rhopilema esculentum Kishinouye 1891 (Cnidaria: Scyphozoa) and the phylogenetic relationship in the related species. Mitochondrial DNA Part B. 2(1):167–168.
- Zou H, Zhang J, Li W, Wu S, Wang G. 2012. Mitochondrial genome of the freshwater jellyfish Craspedacusta sowerbyi and phylogenetics of Medusozoa. PLOS One. 7(12):e51465.