Abstract
This study encoded the complete mitochondrial genomic sequence of the little ringed plover Charadrius dubius. The mitochondrial genome has a total length of 16,864 bp, consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a control region. The nucleotide composition was 23.8% T, 31.6% A, 30.8% C, and 13.8% G. This study provides the basic information on the mitogenome of C. dubius and supports the understanding of mitogenomic information and its phylogenetic relationship within Charadriiformes.
The little ringed plover, Charadrius dubius Scopoli, 1786 has a black mask around its face with yellow eye rings and belongs to the family Charadriidae (Carter and Rogers Citation1998). This species is widely distributed from Africa to Eurasia, with its breeding grounds from Europe and India to East Asia, including the Korean Peninsula (Colwell and Haig Citation2019). According to previous studies about the taxonomy of plovers, Charadrius Linnaeus, 1785 were classified into the two existing major clades (i.e. CRD I and CRD II); C. dubius was also included in CRD I groups (Dos Remedios et al. Citation2015). In this study, we sequenced the complete mitochondrial genome of C. dubius and conducted phylogenetic analysis with the related taxa, which enhanced the basic genetic information of the genus Charadrius. We also conducted phylogenetic analysis with related taxa by sequencing the complete mitochondrial genome of this species.
We captured an individual of C. dubius from the Mangeoyong estuary (N 35°52′51.5″, E 126°40′58.9″) in Gunsan-si, Jellabuk-do, South Korea, using a funnel trap on 30 June 2021; this individual was deposited in the Chonnam National University, Gwangju, South Korea (Voucher storage: Chonnam National University; voucher number: MLR-1; the person in charge of collection was DY Lee; email: [email protected]). The blood sample was collected from the capillary vessel of the captured bird using a micro syringe, and the total genomic DNA was extracted using DNeasy Blood and Tissue kit (Qiagen, Valencia, CA) following the protocol of the manufacturer. Blood and DNA samples were stored in a freezer at −20 °C.
The complete mitochondrial genome of C. dubius was sequenced using Illumina NovaSeq 6000 (Macrogen, Inc., Seoul, South Korea). A total of 5,786,919,729 read bases of filtered data were analyzed to generate 38,416,380 reads of sequence; these were then assembled in Geneious Prime (Kearse et al. Citation2012). Gene annotation was accomplished and circularity was checked using the MITOS web server (Bernt et al. Citation2013, http://mitos.bioinf.uni-leipzig.de/), and secondary structures of tRNA genes were analyzed by comparing them to the nucleotide sequences of other bird species’ tRNA sequences.
The complete mitochondrial genome sequence of C. dubius has a total length of 16,864 bp, which was close to the other reported mitogenomes sizes of Charadrii, which range from 16,791 bp to 17,378 bp; minimal length variation was observed in PCGs, tRNAs, and rRNAs (Chen et al. Citation2018). The mitogenome contains 23.8% T, 31.6% A, 30.8% C, and 13.8% G, which showed a high A + T content; overall AT content of C. dubius mitogenome was 55.3%, which is consistent with previous Charadrii mitogenome studies (Li et al. Citation2014; Hu et al. Citation2017; Chen et al. Citation2018). All protein coding genes have ATN as their start codon, except COX1 and ND5, which use the start codon GTG. The stop codon T– only appeared in the COXIII, ND2, and ND4. A truncated stop codon may be completed through the poly-adenylation of the 3′-end of the mRNA via post-transcriptional processes (Lavrov et al. Citation2002; Chen et al. Citation2018). Additionally, an extra nucleotide (C: cytosine) was present at the position 174 of the ND3 gene for 16 species of Charadrii (Mindell et al. Citation1998; Chen et al. Citation2018); the full length of the D-loop region is 1301 bp.
After removing the termination codon and indels, total length 11,397 bp of the protein-coding genes was used for phylogenetic analysis of 13 species. The tree was constructed with maximum-likelihood method using RAxML version 8.1.2 (Stamatakis Citation2014) with 1000 bootstrap replications. Haematopus ater Vieillot, 1825 and H. ostralegus Linnaeus, 1758 from the family Haematopodidae were used as the outgroup species in this study. This analysis facilitated the construction of a robust phylogenetic tree with high supports for all nodes (see ).
Figure 1. Phylogenetic tree of C. dubius (blue text) with eight other species in Charadriidae, two species in Recurvirostridae, and two species in Haematopodidae. This tree is based on 13 protein-coding genes, constructed using the maximum-likelihood (ML) method. Numbers on each branch indicate the bootstrap support value for 1000 replicates.
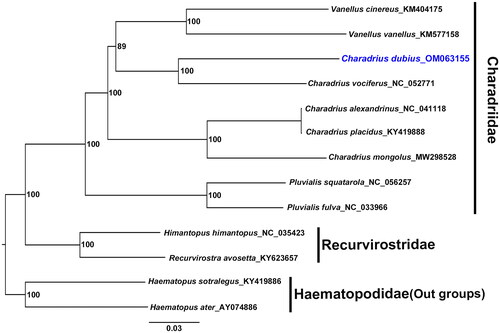
Phylogenetic analysis showed that family Charadriidae is a monophyly group, but the genus Charadrius is not. Moreover, C. dubius was found to be more closely related to C. vociferous Linnaeus, 1758, Vanellus vanellus (Linnaeus, 1758), and V. cinereus (Blyth, 1842) than it was to C. placidus Gray & Gray, 1863, which was previously hypothesized to be its closest relative on the molecular phylogenetic tree (Dos Remedios et al. Citation2015; Colwell and Haig Citation2019). Further research is thus needed to thoroughly understand the morphological characters and conduct in-depth molecular phylogenetic analyses of Charadrius and Vanellus genera.
The results of this study present the complete genetic information of the little ringed plover and propose avenues for future research to improve scientific understanding of the phylogenetic relationships within various species of plovers.
Ethical approval
Permission for the capture and collection of wild animals was granted by the Gunsan City Environment Policy Division [No.-15746 (2021.06.29) 2021-6]. The study species used in this research are not included in the IUCN red list and individuals were not collected from protected areas. All procedures conducted to produce and publish this article were conducted in compliance with the regulations of the Honam National institute of Biological Resources (HNIBR).
Author contributions
WY Kim, HC Sung, JH Lee, and DY Lee conceived the research idea and designed the experiments. SJ Roh, WY Kim, and DY Lee wrote the manuscript. SH Kim got the funding and revised the manuscript. SJ Roh, TW Jung, DJ Lee, and HK Kim conducted laboratory works and analysis. JH Jung, SY Cho, YJ Kim, and JW Kook conducted collection. All authors revised and approved the final manuscript. All authors agree to be accountable for all aspects of the work.
Disclosure statement
The authors report that there are no conflicts of interest to declare that are pertinent to this study. The authors of this study are solely responsible for this paper.
Data availability statement
Genome sequence data that support the findings of this study are openly available from the NCBI GenBank at https://www.ncbi.nlm.nih.gov/, under accession no. OM063155. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA803930, SRP358595, and SAMN25690482, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler P. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Carter M, Rogers D. 1998. Little ringed plover Charadrius dubius: a Kimberley record with comments on morphological and vocal aspects of eastern populations. Aust Bird Watcher. 17(6):269–277.
- Chen W, Zhang C, Pan T, Liu W, Li K, Hu C, Chang Q. 2018. The mitochondrial genome of the Kentish Plover Charadrius alexandrinus (Charadriiformes: Charadriidae) and phylogenetic analysis of Charadrii. Genes Genomics. 40(9):955–963.
- Colwell MA, Haig SM. 2019. The population ecology and conservation of Charadrius plovers. Boca Raton, FL: CRC Press.
- Dos Remedios N, Lee PL, Burke T, Székely T, Küpper C. 2015. North or south? Phylogenetic and biogeographic origins of a globally distributed avian clade. Mol Phylogenet Evol. 89:151–159.
- Hu C, Zhang C, Lei S, Yi Z, Xie W, Zhang B, Chang Q. 2017. The mitochondrial genome of pin-tailed snipe Gallinago stenura, and its implications for the phylogeny of Charadriiformes. P LoS ONE 12:e0175244.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lavrov DV, Boore JL, Brown WM. 2002. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: duplication and nonrandom loss. Mol Biol Evol. 19(2):163–169.
- Li X, Huang Y, Lei F. 2014. Comparative mitochondrial genomics and phylogenetic relationships of the Crossoptilon species (Phasianidae, Galliformes). BMC Genom 16:1–12.
- Mindell DP, Sorenson MD, Dimcheff DE. 1998. An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol. 15(11):1568–1571.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
