Abstract
The genus Hybos Meigen, 1803 belongs to the subfamily Hybotinae of the family Empididae. Here we report a mitogenome of Hybos grossipes (Linnaeus, 1767) as the new representative of the subfamily Hybotinae. The complete mitogenome is 16,325 bp in total, consisting of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a control region. The nucleotide composition is biased toward A and T, accounting for 77.2% of the total. All PCGs start with ATN codons except COI, NAD1, and NAD5, and end with TAA or incomplete stop codon T. The phylogenetic result generated by IQ-Tree based on 13 PGCs showed that the subfamily Hybotinae is monophyletic, and the subfamily Hybotinae is a sister group of the subfamily Ocydromiinae.
Empididae is one of the largest groups in Brachycera (Diptera) with about 180 genera and over 5000 known species (Yang et al. Citation2007). They usually prey on crop pests such as aphids, psyllids, and agromyzid flies, but also on sanitary pests such as mosquitos and black flies. They are very sensitive to environmental changes and have been widely used as an indicator of the quality of the environment and biodiversity (Yang and Yang Citation2004). Members of Hybotinae are characterized by the following features: male and female eyes contiguous on frons; proboscis usually heavily sclerotized and projected forward; prosternum free from proepisternum; anal lobe well developed; two veins arising from cell dm (Sinclair and Cumming Citation2006). Hybos Meigen, 1803 is the largest genus in Hybotinae with over 230 known species distributed all over the world. Twenty-four species are known from the Palearctic Region and 196 species from the Oriental Region (Yang et al. Citation2007; Li et al. Citation2017; Cao et al. Citation2018; Li et al. Citation2022).
The adult specimens of H. grossipes used in this study were collected from Zhubalong (99°01′E, 29°77′N), Batang, Sichuan by Liang Wang on 3 July 2020, and then identified by Ding Yang through the morphological characters (arista short pubescent, R and M divergent apically, hind coxa with 2 thick anterior bristles at extreme tip; hypandrium furcate apically; left surstylus with a fingerlike inner process) (Yang and Yang Citation2004). The specimens were preserved in 95% ethanol and stored in a −20 °C refrigerator in the Entomological Museum of China Agricultural University (CAU), Beijing (Voucher number: CAUlml202203010001, Liang Wang, [email protected]).
The total DNA was extracted from the adults’ muscle tissue using DNeasy Blood & Tissue Kit (Qiagen, Germany). The mitogenome was sequenced on an Illumina NovaSeq 6000 by Novogene Co., Ltd. Raw read data was filtered and trimmed in Trimmomatic v0.30 (Bolger et al. Citation2014). About 6 GB of high-quality data was used to assemble mitochondrial genome with the de novo assembler IDBA-UD (Peng et al. Citation2012). The bait sequence COI was amplified by standard PCR reactions. The primer sequences used for the bait sequence COI amplification were (5′ to 3′): forward primer (ATTCAACCAATCATAAAGATATTGG) and reverse primer (TAAACTTCTGGATGTCCAAAAAATCA). The PCR amplification conditions were as follows: an initial denaturing cycle at 94 °C for 5 min, followed by 35 amplification cycles (denaturing at 94 °C for 30 s, annealing at 44 °C for 30 s and extension at 72 °C for 45 s), and a final extension at 72 °C for 7 min. And the bait was used in BLAST search to locate the target mitogenome, which was carried out with BioEdit 7.0.5.3. The complete mitochondrial genome sequence was annotated by MITOS (Bernt et al. Citation2013) and checked manually in Geneious v.9.0.2 (Kearse et al. Citation2012). The tRNA genes were identified by MITOS and rechecked by the tRNAscan-SE web server (Schattner et al. Citation2005).
The complete mitochondrial genome of H. grossipes (GenBank accession: OM937934) is 16,325 bp in length. It contains 13 typical protein-coding genes, 22 tRNA genes, 2 rRNA genes (12S rRNA and 16S rRNA), and a control region. All genes have similar locations and strands to that of other published dipteran flies (Liu et al. Citation2020; Pi et al. Citation2021; Wang et al. Citation2021; Zhang et al. Citation2021). The nucleotide composition was 39.8% of A, 37.4% of T, 9.0% of G, and 13.8% of C. Among the protein-coding genes, six genes took the start codon of ATG (COX2, COX3, ATP6, NAD4, NAD4L, and COB), four genes used ATT (ATP8, NAD2, NAD3, NAD6) as the start codon, while COX1 gene and NAD1 gene initiated with codon TCG and TTG, respectively. And particularly, NAD5 initiated with codon GTG. Most of the protein-coding genes ended with the conservative stop codon TAA except for that two genes, COX1 and NAD1, which terminated with an incomplete stop codon T + tRNA.
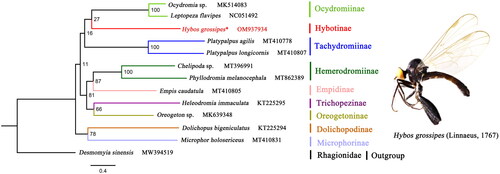
Together with mitogenome data of the other 12 Diptera species retrieved from NCBI (the Accession Numbers are indicated in ), the phylogenetic analysis based on all protein-coding genes of the 13 Diptera species was performed by IQ-TREE web server (Trifinopoulos et al. Citation2016). Desmomyia sinensis was chosen as the outgroup. The topology and nodal support values are shown in . The maximum likelihood (ML) analysis strongly supported the monophyly of the subfamily Hybotinae. The phylogenetic relationship among the seven subfamilies of Empididae is very clear: ((Ocydromiinae + Hybotinae) + Tachydromiinae) + ((Hemerodromiinae + Empidinae) + (Trichopezinae + Oreogetoninae)). Subfamily Hybotinae is the sister group of subfamily Ocydromiinae. The mitogenome of H. grossipes may provide essential DNA molecular data for further phylogenetic and evolutionary studies of the family Empididae.
Ethical approval
The specimen collection protocol was approved by the Ethics Committee of China Agricultural University. The studies did not involve endangered or protected species.
Authors’ contributions
MLL, XL, YTG, XDC, and DY contributed to the conception and design of the research. MLL, YTG, and XDC performed experiments and analyzed the data. MLL drafted the manuscript, and XL and DY revised the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s). All authors have approved the manuscript for publication and agreed to be accountable for all aspects of the work.
Data availability statement
Mitochondrial genome sequence can be accessed via accession number OM937934 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, Bio-Sample and SRA, numbers are PRJNA811344, SAMN26319898, and SRR19632878, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Cao YK, Yu H, Wang N, Yang D. 2018. Hybos Meigen (Diptera: Empididae) from Wangdongyang Nature Reserve, Zhejiang with descriptions of three new species. Trans Am Entomol Soc. 144(1):197–218.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li ML, Fatima N, Lin C, Yang D. 2022. Four new species of Hybos (Diptera: Empididae) from Gaoligongshan, China. Entomotaxonomia. 44(2): 134–143.
- Li XL, Wang N, Yang D. 2017. Hybos Meigen (Diptera: Empididae) from Wanglang National Nature Reserve, Sichuan. Trans Am Entomol Soc. 143(2):435–452.
- Liu Y, Wang MQ, Chen NZ, Yang D. 2020. The mitochondrial genome of Leptopeza flavipes (Diptera: Empididae). Mitochondrial DNA Part B. 5(3):2942–2943.
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428.
- Pi Z, Jiang Y, Wang S, Cai J, Guo Y. 2021. The complete mitochondrial genome of Neomyia cornicina (Diptera: Muscidae). Mitochondrial DNA Part B. 6(9):2691–2692.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Sinclair BJ, Cumming JM. 2006. The morphology, higher-level phylogeny and classification of the Empidoidea (Diptera). Zootaxa. 1180(1):1–172.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Wang J, Li X, Du R, Liu Y. 2021. The complete mitogenome of Chlorops oryzae Matsumura (Diptera: Chloropidae). Mitochondrial DNA B Resour. 6(7):1844–1846.
- Yang D, Yang CK. 2004. Fauna Sinica Insecta (Vol. 34) Diptera Empididae. Beijing (China): Science Press.
- Yang D, Zhang K, Yao G, Zhang J. 2007. World catalog of Empididae (Insecta: Diptera). Beijing (China): Agricultural University Press.
- Zhang B, Gao S, Yang D. 2021. The mitochondrial genome of Epiphragma (Epiphragma) mediale (Diptera: Limoniidae). Mitochondrial DNA Part B. 6(4):1321–1323.