Abstract
Primula odontocalyx (Franch.) Pax (1905) is a perennial herb of the genus Primula in Primulaceae with heterostyly and ornamental value. Here, the chloroplast genome of P. odontocalyx was sequenced, assembled, and annotated. The complete chloroplast genome was a closed-circular molecule of 151,738 bp in length, containing a large single-copy region (LSC) of 83,817 bp, a small single-copy region (SSC) of 17,529 bp, and two inverted repeat (IR) regions of 25,196 bp. A total of 115 unique genes were annotated in the whole cp genome, including 81 protein-coding genes, 4 rRNA genes, and 30 tRNA genes. Phylogenetic analysis confirmed the close relationship between P. odontocalyx and Primula moupinensis, and both species belong to Sect. Petiolares Pax.
Primula odontocalyx (Franch.) Pax (1905) is a perennial herb () of Sect. Petiolares Pax in the genus Primula (Primulaceae) (Hu and Kelso Citation1996). Primula odontocalyx was placed in Sect. Davidii by Smith and Fletcher (1944). But now it was placed in Sect. Petiolares due to the absence of long hairs and basal bud scales at an early stage as shown by most specimens of this species (Richards Citation2003). Primula odontocalyx is mainly distributed in thickets and forest margins at an altitude of 900–3400 m in southern Gansu, western Hubei, southern Shanxi, Henan, and Sichuan Provinces of China (Hu and Kelso Citation1996). The flower of P. odontocalyx has 1–3 or rarely as many as 8 pink or lilac-rose corollas (Hu and Kelso Citation1996), this species, therefore, exhibits high ornamental value. In this study, we reported the first chloroplast genome of P. odontocalyx, which is useful for its taxonomic, systematics, and evolutionary studies.
Figure 1. Primula odontocalyx. This image is provided by the corresponding author Yuan Huang, and is used with permission.

Fresh leaf of P. odontocalyx was collected from Chengkou of Sichuan Province, China (31.948°N, 108.665°E), and the corresponding voucher specimens were deposited in the Herbarium of Yunnan Normal University (Kunming, China; Jianlin Hang, [email protected]) under the accession number of HY-24. Total genomic DNA was extracted using a modified CTAB method (Porebski et al. Citation1997) and 300 bp short-insert libraries were constructed following the manufacturer’s protocol (Illumina Inc., USA), and then paired-end sequenced on Illumina Hiseq X Ten sequencer platform. We obtained 14,914,454 filtered reads and then assembled the plastid genome using software NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017) with Primula sinensis Sabine ex Lindley as reference genome (GenBank accession No: NC_030609). The assembled genome was annotated using Geneious v2020.1.1 (Kearse et al. Citation2012), with Primula moupinensis (Genbank accession No. NC_050244) as the reference genome.
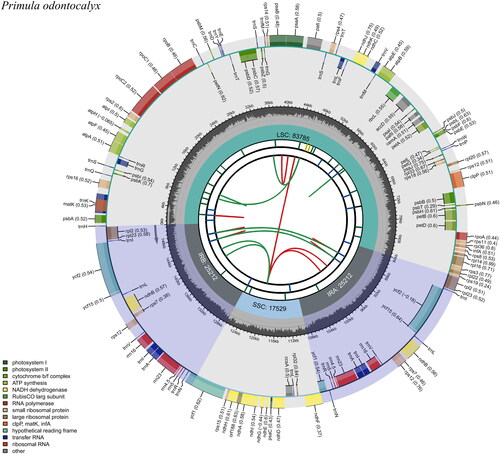
The complete plastid genome of Primula chionantha was assembled into a circular molecule with a length of 151,738 bp (), average coverage of 506.5, and GC content of 37.0%, respectively, the NCBI accession number of the plastid genome is ON416872. The plastid genome of this species has a large single-copy region of 83,817 bp and a small single-copy region of 17,529 bp, which are separated by a pair of inverted repeats of 25,196 bp. In total, the plastid genome has 134 genes, 89 protein-coding genes (CDS), 37 tRNA genes, and 8 rRNA genes are annotated in the plastome, of which 115 genes, 81 protein-coding genes, 30 tRNA genes, and 4 rRNA genes are unique, respectively.
Figure 2. Genomic map of Primula odontocalyx chloroplast genome generated by CPGview. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise. Genes belonging to different functional groups are color-coded. The dashed area in the inner circle indicates the GC content of the chloroplast genome of Primula odontocalyx.
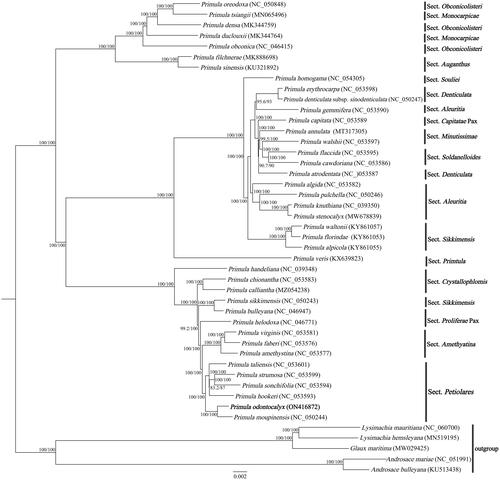
In order to further determine the phylogenetic position of P. odontocalyx within the genus Primula, we downloaded 39 Primula plastomes and two Androsace species, two Lysimachia species, and Glaux maritima (Liu et al. Citation2021) as outgroups. A total of 45 chloroplast genome sequences were aligned using MAFFT v7.47 (Katoh and Standley Citation2013). A maximum-likelihood phylogenetic tree was constructed using IQ-TREE v1.6.10 (Nguyen et al. Citation2015), and the best-fitted model is TVM + F+R2 using ModelFinder according to Bayesian information criterion (Kalyaanamoorthy et al. Citation2017). Branch supports were tested using ultrafast bootstrap (UFBoot) (Hoang et al. Citation2018) and SH-like approximate likelihood ratio test (SH-aLRT) (Guindon et al. Citation2010) with 10,000 replicates. The ML tree showed that P. odontocalyx was highly supported to be a sister species to P. moupinensis, forming a monophyletic clade with the rest species of Sect. Petiolares (). This result supports that P. odontocalyx belongs to Sect. Petiolares, rather than Sect. Davidii. Additionally, the topologic structure of the phylogenetic tree showed that the relationships between Primula, Androsace, Lysimachia, and Glaux are consistent with previous phylogenetic studies (Mast et al. Citation2001).
Ethical approval
This study does not involve ethical issues. The sample of this study is not a protected plant, and specimens are collected in accordance with the laws and regulations of the country where we are located.
Authors’ contributions
Yuan Huang designed the study. Yunqi Liu, Li Zhang, Shubao Wang, and Rui Li collected materials and performed the experiments and analysis. Yunqi Liu drafted the original manuscript.
Supplemental Material
Download JPEG Image (711.2 KB)Supplemental Material
Download JPEG Image (88.1 KB)Supplemental Material
Download JPEG Image (270.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s). All authors revised and approved the manuscript.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov/] under accession no. ON416872. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA834967, SRR19141931, and SAMN28088246, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Hu Q, Kelso S. 1996. Primulaceae. In: Wu CY, Raven PH, editors. Flora of China. Vol. 15. Beijing (China); St. Louis (MO): Science Press and Missouri Botanical Garden; p. 122–123.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu YQ, Chen X, Zhang L, Huang Y. 2021. The complete chloroplast genome of Glaux maritima, a monotypic species. Mitochondrial DNA B Resour. 6(8):2137–2138.
- Mast AR, Kelso S, Richards AJ, Lang DJ, Feller DM, Conti E. 2001. Phylogenetic relationships in Primula L. and related genera (Primulaceae) based on noncoding chloroplast DNA. Int J Plant Sci. 162(6):1381–1400.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15.
- Richards J. 2003. Primula. 2nd ed. London (UK): Batsford; p. 158–163.