Abstract
In this study, the complete mitogenome of Grapsus albolineatus (Lamarck, 1818) (Crustacea: Grapsoidea) was sequenced. The mitogenome of G. albolineatus was a circular molecule with 15,578 bp length. Its nucleotide composition was 26.81% A, 16.37% G, 34.51% T, and 22.31% C. It comprised 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA), and two ribosomal RNA (rRNA). All PCGs were initiated by ATN codons, except for the atp8 and nad1 genes. Ten PCGs used a common stop codon of TAA or TAG, and the other three ended with a truncated stop codon (a single stop nucleotide T). Phylogenetic analysis revealed that G. albolineatus was closely related to species from the genera Pachygrapsus and Metopograpsus.
The tropical rocky shore crab Grapsus albolineatus (Lamarck, 1818) (Crustacea, Decapoda, Grapsoidea), is a crucial species engineering coastal soil by its litter feeding activities. It is widely distributed along the rocky coastlines of China (Guangdong, Hainan, Taiwan), Japan, Hawaii, and Australia (Dai et al. Citation1986). The fluctuating asymmetry for leg segments of G. albolineatus can be applied as a developmental instability approach to identify stressed coastal area. In addition to maintaining ecosystem stability, the larvae of G. albolineatus are used as natural bait for aquaculture (Gao et al. Citation2000). The alcalase hydrolysis of rocky shore crab G. albolineatus can produce bioactive peptides with potent antioxidant and antibacterial activities as affected by the degree of hydrolysis up to a certain level (Shaibani et al. Citation2019). Peptide fractions isolated from the protein hydrolysate of G. albolineatus inhibited the proliferation of cancer cells and can be regarded as new agents for nutraceutical and pharmaceutical applications (Shaibani et al. Citation2020). The interest in the study of G. albolineatus has increased significantly; however, the research on this species is limited at the molecular level. Due to coastal land reclamation and commercial exploitation, the population genetic diversity of G. albolineatus is under threat. Therefore, it is necessary and urgent to perform genetic investigations on G. albolineatus. Thus, the complete mitogenome of G. albolineatus was sequenced in order to provide a theoretical basis for species identification and facilitate the protection of crab populations.
The samples used for sequencing were collected in Sanjiang Town, Haikou City, Hainan Province, China (N18°09′34″, E108°56′30″). The samples were preserved in 100% alcohol when collected, and then stored at −80 °C freezer for long-term storage (Pu et al. Citation2017). The specimen was deposited in the marine specimen repository of Yantai University (YTU-SKY-201900370102, contact person: Jiangyong Qu; email: [email protected]). Genomic DNA was extracted using column animal mtDNAOUT (Baiaolaibo Science Technology Co., Ltd., Beijing, China, BTN120501) and quality control was subsequently carried out on the purified DNA samples. The mitogenome sequence was loaded to an Illumina NovaSeq 6000 sequencing platform (Illumina, San Diego, CA) and assembled using SPAdes v3.10.1. The complete mitogenome was annotated using MITOS webserver (Bernt et al. Citation2013), subsequently improved by Geneious Prime, and phylogenetic tree was constructed by MrBayes 3.2 program (Ronquist et al. Citation2012).
The complete mitogenome of G. albolineatus (GenBank accession number: MT755825) was 15,578 bp. The nucleotide composition of this mitogenome was A = 4175 bp (26.8%), T = 5375 bp (34.5%), C = 3473 bp (22.3%), and G = 2555 bp (16.4%). Previous studies have shown similar findings in Crustacea, the GC content was lower than the AT content (Tsang et al. Citation2015; Song et al. Citation2020). The mitogenome contained 13 protein-coding genes (PCGs), that included seven NADH dehydrogenases (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6), three cytochrome c oxidases (cox1, cox2, and cox3), one cytochrome b (cob), and two ATP synthases (atp6 and atp8). Furthermore, it contained 22 transfer RNA (tRNA) and two ribosomal RNA (rRNA). The total length of the 13 PCGs was 11,163 bp. Seven of the 13 PCGs (nad4, nad4L, nad5, cox1, cox2, cox3, and cob) started with ATG, nad2 and atp6 started with ATT, nad3 started with ATC, and nad6 started with ATA; however, the initiation codon of atp8 and nad1 could not be determined. Eight of the 13 PCGs had the stop codon TAA (nad1, nad3, nad4L, nad5, nad6, atp6, atp8, and cox3), and two had TAG (nad2 and nad4). It should be noted that the other three PCGs (cox1, cox2, and cob) terminated with an incomplete stop codon, which was completed by the addition of 3′ A residues to the mRNA. ‘Truncated or incomplete stop codon’ also exists in Leptestheria brevirostris, Gondwanalimnadia sp., and Upogebia major (Tladi et al. Citation2020; Emami-Khoyi et al. Citation2021; Sun and He Citation2021). Two rRNA genes, 12SrRNA (827 bp) and 16SrRNA (1402 bp) are separated by a trnV-GTA. All tRNAs had a typical cloverleaf structure and their length ranged from 63 to 72 bp.
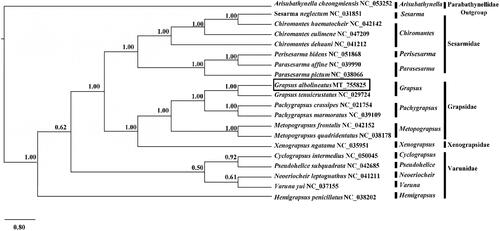
To clarify the phylogenetic position of G. albolineatus, the mitogenome sequences of 18 closely related species and one outgroup species Euphausia superba were downloaded from GenBank database. Thirteen shared PCGs in each of the 20 complete mitochondrial genomes were aligned to generate a phylogenetic tree based on the Bayesian inference (BI) method by Mr. Bayes. Support values for each node were calculated using Bayesian posterior probability (BPP). The results confirmed that G. albolineatus was clustered with G. tenuicrustatus and was rooted in other Grapsoidea species (). The genus Grapsus was most closely related to the genera Pachygrapsus and Metopograpsus, both of which belong to the family Grapsidae, which have not been previously reported. Altogether, our results provided insights into the crustacean mitochondrial genome diversity and the evolution of decapods.
Ethics statement
All animal protocols have been reviewed and approved by the experimental animal welfare and ethics review committee of Yantai University.
Author contributions
Jiangyong Qu, Xumin Wang, and Lijun Wang conceived the original idea; Xiyue Wang, Shuyi Xu, and Lijun Wang analyzed the data and wrote the manuscript; Wenwen Chen, Xintong Hu, Lingxue Cui, and Xue Li performed the samples collection, DNA extractions and helped to the acquisition, analysis, or interpretation of data for the work.
Acknowledgements
The authors thank Chuanlin Liu for the help on the sample collection and identification and Shanghai Biozeron Biotech. Co., Ltd for sequencing work.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
All data in this study are openly available in GenBank (accession number MT755825). Original data submitted in NCBI, BioProject: PRJNA785768, SRA: SRR17110818, and BioSample: SAMN23576620.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dai AY, Yang SL, Song YZ, Chen GX. 1986. Chinese marine crabs. Beijing: Ocean Press.
- Emami-Khoyi A, Tladi M, Dalu T, Teske PR, Jansen van Vuuren B, Rogers DC, Nyamukondiwa C, Wasserman RJ. 2021. The complete mitogenome of Leptestheria brevirostris Barnard, 1924, a rock pool clam shrimp (Branchiopoda: Spinicaudata) from Central District, Botswana. Mitochondrial DNA B Resour. 6(2):608–610.
- Gao TX, Zhang XM, Atanabe A, Jiao Y. 2000. Genetic relationship among seven Grapsidae species. Acta Hydrobiol Sin. 24(6):621–629.
- Pu DQ, Liu HL, Gong YY, Ji PC, Li YJ, Mou FS, Wei SJ. 2017. Mitochondrial genomes of the hoverflies Episyrphus balteatus and Eupeodes corollae (Diptera: Syrphidae), with a phylogenetic analysis of Muscomorpha. Sci Rep. 7:44300.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shaibani ME, Heidari B, Khodabandeh S. 2020. Antioxidant and antibacterial properties of protein hydrolysate from rocky shore crab, Grapsus albolineatus, as affected by progress of hydrolysis. Int J Aquat Biol. 8(3):184–193.
- Shaibani ME, Heidari B, Khodabandeh S, Shahangian S. 2019. Isolation of bioactive peptides from rocky shore crab, Grapsus albolineatus, protein hydrolysate with cytotoxic activity against 4T1 cell line. Ecopersia. 7(3):175–181.
- Song J, Chen P, Tian M, Ji N, Cai Y, Shen X. 2020. The first mitochondrial genome of Fistulobalanus albicostatus (Crustacea: Maxillopoda: Sessilia) and phylogenetic consideration within the superfamily Balanoidea. Mitochondrial DNA Part B. 5(3):2776–2778.
- Sun X, He J. 2021. The complete mitochondrial genome and phylogenetic analysis of Upogebia major (De Haan, 1841). Mitochondrial DNA Part B. 6(3):970–971.
- Tladi M, Dalu T, Rogers DC, Nyamukondiwa C, Emami-Khoyi A, Oliver JC, Teske PR, Wasserman RJ. 2020. The complete mitogenome of an undescribed clam shrimp of the genus Gondwanalimnadia (Branchiopoda: Spinicaudata), from a temporary wetland in Central District, Botswana. Mitochondrial DNA Part B. 5(2):1238–1240.
- Tsang LM, Shen X, Chu KH, Chan BKK. 2015. Complete mitochondrial genome of the acorn barnacle Striatobalanus amaryllis (Crustacea: Maxillopoda): the first representative from Archaeobalanidae. Mitochondrial DNA. 26(5):761–762.