Abstract
The first complete mitochondrial genome sequencing of Asiatic Water snake or Checkered Keelback or Fowlea piscator (=Xenochrophis piscator) was carried out using Next-Generation Sequencing technology. The complete mitochondrial genome of Asiatic Water snake is 16,999bp long with a base composition of 33% A, 28% T, 12% G and 27% C, with a GC content of 39%. Like the typical snake mitochondrial genome, F. piscator also shows relatively similar mitogenome arrangement comprising 37 genes including 13 protein-coding genes, 22 tRNA genes, two rRNA genes and two non-coding regions or a duplicated control region (D-Loop) along with an origin of replication. Nine genes including eight tRNAs and NAD6 were encoded on the Light or L-strand. Phylogenetic analyses using the complete mitochondrial genome of F. piscator demonstrate a close relationship with the family Colubridae and sub family Natricinae.
Introduction
The Asiatic water snake or Checkered keelback or Fowlea piscator (=Xenochrophis piscator) (Schneider 1799) are non-venomous, almost 3–5 ft long, fleshy snakes found near freshwater ponds, lakes or rivers. They are listed in schedule III of CITES and play an important role in controlling ecological balance (Das Citation2000). They primarily feed on small fishes, frogs, and insects (Whitaker and Dattatri Citation1986; Hossain Citation2016). They are rich in protein content and hence used as food items in East Asian countries. Despite their wide distribution across Asian countries, the species is presently declining due to habitat loss caused by rapid urbanization, illegal trafficking to East Asian countries for meat and indiscriminate killing of snakes (Das Citation2013).
There are presently eight species in the genus Fowlea (Schneider 1792; Vogel and David Citation2012). Most studies on mitochondrial DNA are restricted to the analysis of Cytochrome b, COI, 12S rRNA, 16S rRNA and NADH2. But, there are no reports for the characterization and evaluation of the complete mitochondrial genome of any species from the genus Fowlea. Here, we have sequenced and characterized for the first time the Complete Mitochondrial Genome of Fowlea piscator from Eastern India.
Fowlea piscator’s biological sample was collected from the Zoological garden, Alipore, Kolkata, West Bengal, India (Latitude 22° 32'and longitude 88°24′). The sample was stored in the repository of Central Forensic Science Laboratory, Kolkata, West Bengal, India (https://cfslkol.in; Dr. Soma Roy; [email protected]) under voucher no. CFSLK_IM_XP512567. The extraction of genomic DNA was carried out using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. For performing NGS, 150 bp paired-end libraries were prepared, tagged and run on Illumina NextSeq 500 platform, yielding 4,200,774 raw reads. The mtDNA sequences thus generated were compared with the Tiger Keelback or Rhabdopis tigrinus as RefSeq from NCBI (GenBank Accession No. KU641019.1) and assembled using MITObim version1.9 (Hahn et al. Citation2013). Annotations were generated using MITOS WebServer 2. (Bernt et al. Citation2013). 22tRNAs were analyzed using tRNAscan-SE v.2.0 (Lowe and Chan Citation2016). The phylogenetic relationship was established using MEGA 7 (Kumar et al. Citation2016) which retrieved the complete mitochondrial genome of 24 species from Colubridae, Pythonidae and closely related family Crocodilidae as an outgroup from Genbank.
The total length of F. piscator is 16,999 bp (GenBank Accession No.: OK110208.1) with a base composition of 33% ‘A,’ 28% ‘T,’ 12% ‘G,’ and 27% ‘C,’ with a ‘GC’ content of 39%. The 17 kb complete mitochondrial genome contained 22 transfer RNA (tRNA) genes, 13 protein-coding genes (PCGs), two ribosomal RNA (12S rRNA and 16S rRNA) genes, two control regions (CRs), and one origin of light-strand replication (OL), which is similar to typical snake mtDNA (Dong and Kumazawa Citation2005; Dubey et al. Citation2012; Xu et al. Citation2015; Sun et al. Citation2017; Wu et al. Citation2017; Liang et al. Citation2019; Yu et al. 2019). Among all thirteen (13) PCGs, twelve (12) were encoded on + ve or H-strand, and one NAD6 gene was encoded on the –ve or L-strand. Eight (8) PCGs started with ‘ATG’; NAD2 with ‘ATT,’ NAD3 & NAD5 with ‘ATC,’ COX1 with ‘ATA’ and NAD1 with ‘ACA.’ Eight (8) PCGs terminated with the complete stop codon ‘TAA’ i.e., Cytb, COX2, ND1, ND4L, ND4, ND5, ATP8 and ATP6, whereas, COX1 terminated with ‘AGG’ as a stop codon, and the rest showed an incomplete stop codon ‘T––.’ Furthermore, the length of the 22 tRNA genes ranged from 57 bp (tRNA-Iso) to 74 bp (tRNA-Tyr). The 12 s rRNA and 16 s rRNA genes were 927 bp and 1471 bp long respectively and were separated by the tRNA-Val gene. As per the annotation, the OL was located between tRNA-Asn and tRNA-Cys genes with a length of 39 bp.
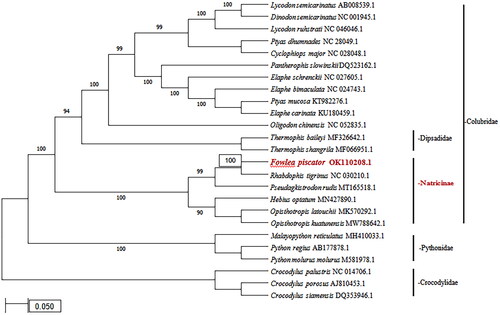
The phylogenetic tree was constructed and analyzed in MEGA 7.0 (Kumar et al. Citation2016) by using complete mitochondrial genome sequencing of F. piscator and 24 other species of which 18 species were from the family Colubridae, and 6 from distant or outgroup family Pythonidae and Crocodilidae. The Maximum Likelihood tree showed that F. piscator clustered with the Rhabdophis tigrinus and Pseudagkistrodon rudis, and then clustered with Hebius optimum, Opisthotropis latouchii and Opisthotropis kuatunensis which are mainly members of sub family Natricinae under family Colubridae (). The mitogenome acquired in this study can be helpful in different population genetics studies or evolutionary studies of these species and provide genomic resources for further studies on Natricinae as well as on Colubridae.
Figure 1. The Phylogenetic tree was constructed through MEGA 7.0 (Kumar et al. Citation2016) using the Maximum Likelihood method based on the Tamura-Nei model involving 25 complete mitochondrial genome sequences of Colubridae, Pythonidae and Crocodylidae species along with their accession numbers. All positions, containing gaps and missing data were eliminated. Bootstrap values are shown for each nodes based on 1000 replicates.
Ethical approval
Permission for the collection of biological samples had been obtained from competent authority: under Section 12 of Wildlife Protection Act, 1972, Ministry of Environment and Forests (Wildlife Divisions), Govt. of India, New Delhi, India Ref: F. No. 1-28/20015 WL-I Dated: 21st May, 2015 and Principal Conservator and Chief Wildlife Warden, West Bengal, India. Memo No. 3843/WL/4R-6/2015; dated 13th July, 2017.
Author contributions
All the authors have contributed to the collection of literature and reflected their work experience on wildlife case handling in this original article. I. M and I. H conceptualized the outline of the work. Sample collection was done by I.M and D.D. Laboratory experiments, analysis of data and drafting of the manuscript was done by I. M. Both S. R and I. H simultaneously contributed in data analysis and manuscript editing along with I M.
Acknowledgements
The authors would sincerely like to acknowledge the Directorate of Forensic Science Services for giving the opportunity to work on Wildlife Forensics at Central Forensic Science Laboratory, Kolkata, West Bengal and Chief Principal Conservator cum Chief Wildlife Warden, Kolkata, West Bengal, India to allow us for snake sample collections from Zoological Garden, Alipore, Kolkata, West Bengal, India.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete mitogenome sequence data that support the findings of this study are available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/nuccore/OK110208.1) under the accession no. OK110208.1. The associated BioProject, SRA and Bio-Sample numbers are PRJNA818295, SRR19137827 and SAMN27403394 respectively.
References
- Bernt M, Donath A, J€uhling F, Externbrink F, Florentz C, Fritzsch G, P€utz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Das CS. 2000. Environmental impact assessment of snake in Sundarban, West Bengal. Indian J Landsc Syst Ecol Stud. 23(1):123–135.
- Das CS. 2013. Declining snake population—why and how: a case study in the Mangrove Swamps of Sundarban, India. Eur J Wildl Res. 59(2):227–235.
- Dong S, Kumazawa Y. 2005. Complete mitochondrial DNA sequences of six snakes: phylogenetic relationships and molecular evolution of genomic features. J Mol Evol. 61(1):12–22.
- Dubey B, Meganathan PR, Haque I. 2012. Complete mitochondrial genome sequence from an endangered Indian snake, Python molurus molurus (Serpentes, Pythonidae). Mol Biol Rep. 39(7):7403–7412.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–abaiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Hossain M. 2016. Food habits of Checkered Keelback, Xenochrophis piscator (Schneider, 1799), in Bangladesh. Bangladesh J Zool. 44(1):153–161.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liang Z, Haiying J, Linmiao L, Jinping C. 2019. Complete mitochondrial genome of Opisthotropis andersonii (Serpentes: Colubridae) determined using next-generation sequencing. Mitochondrial DNA B. 4(1):1012–1013.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: search and contextual analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Sun HJ, Li E, Sun L, Yan P, Xue H, Zhang F, Wu XB. 2017. The complete mitochondrial genome of the greater green snake Cyclophiops major (Reptilia, Serpentes, Colubridae). Mitochondrial DNA B Resour. 2(1):309–310.
- Vogel G, David P. 2012. A revision of the species group of Xenochrophis piscator (Schneider, 1799) (Squamata: Natricidae) (Zootaxa 3473) 60 pp.
- Whitaker R, Dattatri S. 1986. The role of reptiles in controlling food pests. Hamadryad. 11(1&2):23–34.
- Wu W, Jiang D, Sun F. 2017. Next-generation sequencing yields the complete mitochondrial genome of the Shangrila hot-spring snakes (Thermophis shangrila; Reptilia: Colubridae). Mitochondrial DNA B Resour. 2(1):327–328.
- Xu CZ, Zhao S, Han XM. 2015. Sequence and organization of the complete mitochondrial genome of Hebius vibaka ri ruthveni from China. Mitochondrial DNA. 27:1–2.
- Yu W, Pan L, Hui L, Chen S. 2019. The complete mitochondrial genome of Opisthotropis latouchii (Squamata: Colubridae). Mitochondrial DNA B. 4(1):1437–1438.
