Abstract
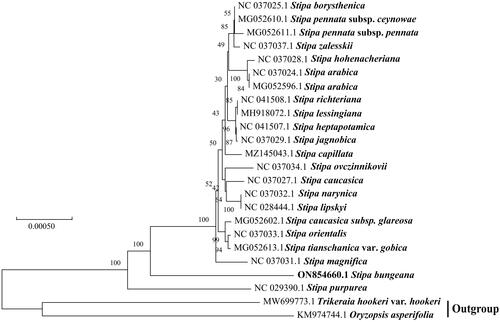
Stipa bungeana Trin. 1833 is an important forage grass in Poaceae, widely distributed in the temperate steppe of Northern China, with strong grazing tolerance and feeding value. In this study, we performed the complete chloroplast (cp) genome sequence of S. bungeana to explore its phylogenetic position with other Stipa. The results showed that the circular complete cp genome of S. bungeana was 137,759 bp in length, including a large single copy (LSC) of 81,652 bp, a small single copy (SSC) of 12,817 bp, and two inverted repeats (IR) of 21,645 bp. The GC content accounts for 43.71% and annotated 134 single genes, which include 87 protein-coding genes, eight rRNA genes, and 39 tRNA genes. Maximum-likelihood (ML) phylogenetic tree suggested that the S. bungeana was closely related to other Stipa except for S. purpurea.
Stipa bungeana Trin. 1833, is an important forage grass of Stipa in the family Poaceae, widely distributed in the temperate steppe of northern China, and it also is a typical representative species of the Eurasian steppe (Kuo Citation1987). The main difference between S. bungeana and other Stipa is that the awn needle of S. bungeana is 3–5 cm long, slightly curved, and shiny as hair, 2-geniculate, column 1–1.5 cm to first bend, 0.5–1.0 cm to second bend (Kuo and Sun Citation1982; ). S. bungeana is a typical bottom grass, with the characteristics of trampling resistance, suitable for use in grazing land, this species germinates earlier in spring and has stronger tillering ability, it is rich in nitrogen-free extract and crude fiber, the cattle and sheep are more fond of feeding and tend to gain weight (Kuo Citation1987; Jia Citation1997). To assess the relationship between genetic diversity and geographical distribution, previous studies have reported that the genetic diversity of S. bungeana is higher at the species level than at the population level, and which is no significant relationship between the genetic distance and geographical distance (Jing et al. Citation2013). The wild Stipa species in the natural ecosystem are prone to hybridization, the studies have found that the offspring of this natural hybridization had a great variation in morphology, and the taxonomic status of S. bungeana is not certain from the genome, it should belong to the section Leiostipa, but from the perspective of genetics, it is closer to S. breviflora of the section Barbatae (Baiakhmetov et al. Citation2020). A study of the hybrid-complex for S. heptapotamica showed that the hybridization between the wild Stipa each other of Poaceae is an introgression event (Nobis et al. Citation2019). However, the uncertainty of the taxonomic status of S. bungeana (Baiakhmetov et al. Citation2020) and the evidence for the chloroplast (cp) whole genome and its phylogenetic status still need to explore. Angiosperm Phylogeny Group (APG) classification system facilitates our understanding of plant evolution and classification (Chase et al. Citation2016), such as gene markers. The gene fragments (accD, matk, rps16-trnQ, psbA-trnH, etc) in plastids are commonly used to mark phylogenetic status and taxonomic attributes of species (Mishra et al. Citation2016; Van Do et al. Citation2021). It has been reported that these barcodes can be used as a species identification tool for Stipa of Poaceae, such as S. pennata (Krawczyk et al. Citation2018), S. capillata (Baiakhmetov et al. Citation2021), and S. lipskyi (Myszczyński et al. Citation2016). These highlight the importance of cp genomes in taxonomy. Thus, we determined the complete cp genome of S. bungeana to analyze and confirm a phylogenic position (Nobis et al. Citation2019), as well as to provide useful information for further studies.
Fresh leaves of S. bungeana were sampled from desert steppe in eastern and western Ningxia, China (38°7′38.57 E, 106°34′27.31 N, alt. 1152 m), and samples are stored in dry ice buckets. The voucher specimen (2022-Stipa_bungeana001) was deposited at the herbarium of the Institute of Forestry and Grassland Ecology, Ningxia Academy of Agriculture and Forestry Science (http://www.nxaas.com.cn/, Wangsuo Liu, email: [email protected]). Total genomic DNA was extracted according to the modified CTAB method of Stefanova et al. (Citation2013), and stored in the −80 °C refrigerator in the laboratory. The genome sequencing was conducted by Illumina Hiseq 2500 at Biomarker Technologies Corporation. 25,873,239 paired-end reads were obtained and 20,893,700 reads were used to assemble into reference sequences ranging in length from 50 to 151 bp after trimming. The high-quality sequences were assembled by the assembler SPAdes3.11.0 (Nurk et al. Citation2013). The annotation was performed by Plann (Huang and Cronk Citation2015). Then, the complete cp genome map was drawn by OGDRAW-Draw Organelle Genome Maps (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Greiner et al. Citation2019). The sequence of S. bungeana complete cp genome has been submitted to the NCBI database (accession number ON854660).
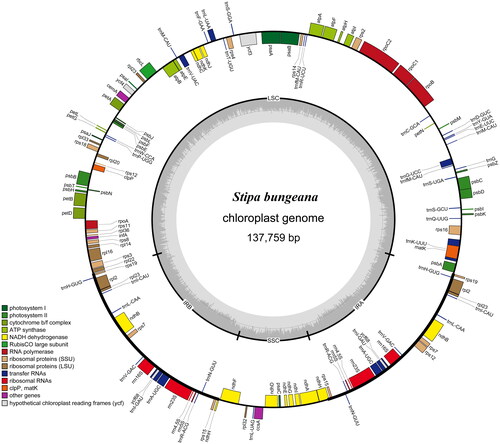
The S. bungeana cp genome is 137,759 bp in length, including two inverted repeat regions (21,645 bp) that are separated by a small single-copy region (12,817 bp) and a large single-copy region (81,652 bp). The cp genome contains 134 single genes, including 87 protein-coding genes (CDS), eight rRNA, and 39 tRNA genes (). The GC content is 43.71%. Most of these genes are single copy, whereas eight CDS (rps19, rps15, rps7, rp123, rp12, ndhH, ycf68, ndhB), 10 tRNAs (trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnM-CAU, trnN-GUU, trnR-ACG, trnV-GAC, trnfM-CAU) and four rRNAs (rrn-16S, rrn-23S, rrn4.5, rrn5) occur as double copies.
Figure 2. Gene map of S. bungeana chloroplast genome. Genes shown inside the circle indicate that the direction of transcription is clockwise, while those shown outside are counterclockwise. Different groups of functional genes are indicated in different colors. The GC content is shown in the dashed area in the inner circle.
The cp genomes of 21 Stipa in Poaceae and two outgroups were downloaded from the NCBI database and aligned with S. bungeana using MAFFT-7.037 (Katoh and Standley Citation2013). A maximum-likelihood tree was constructed by MEGA-X (Kumar et al. Citation2018) based on 24 species (). The phylogenetic tree showed that the S. bungeana clustered into a clade with most species of Stipa except S. purpurea. This change may be related to the higher genetic differentiation value between S. bungeana and other Stipa species, or the event of genetic drift (Jing et al. Citation2013). Krawczyk et al. (Citation2018) analyzed 21 newly sequenced complete plastid genomes from 19 groups of Stipa through the gene fragment analysis, and they found that the multilocus barcodes composed of ndhH, rpl23, ndhF-rpl32, rpl32-ccsA, psbK-psbI and petA-psbJ for Stipa were performed the best, but the effectiveness was less than 70% of the analyzed taxa, indicating that these markers did not apply to all Stipa. Hybridization among species of Stipa in different habitats may have resulted in different highly variable gene segments (Baiakhmetov et al. Citation2020). In the view of the phylogenetic clustering tree, although it was related to other Stipa, it clustered in a single clade (). This clustering may confirm that the existence of introgression in Stipa, which is likely to be closely related to hybridization (Nobis et al. Citation2019). In our study, S. bungeana was distributed in water-deficient dunes, therefore, environmental filtration and interspecific hybridization were inevitable, and perhaps these factors are interesting areas of future research to explore genetic changes in the same species of Stipa. Our study provides ideas for future studies of S. bungeana from the perspective of the cp genome.
Ethical approval
Stipa bungeana is a widely distributed native grass and is not a protected species. Therefore, the collection and research process are conventional and does not need to follow strict standards.
Author contributions
WL and Z-JW planned and designed the research. YT and BJ sampled the plant materials, YT and WL performed experiments, and WL analyzed the data. Z-JW and WL wrote the manuscript.
Acknowledgments
Thanks to Professor Xiaowei Li of Ningxia University for identifying this species.
Disclosure statement
The authors report there are no competing interests to declare.
Data availability statement
The data that support the findings of this study are openly available in the NCBI database at https://www.ncbi.nlm.nih.gov/, reference number ON854660. The associated BioProject, SRA, and Biosample numbers are PRJNA852531, SRR19959630, and SAMN29333018, respectively.
Additional information
Funding
References
- Baiakhmetov E, Guyomar C, Shelest E, Nobis M, Gudkova PD. 2021. The first draft genome of feather grasses using SMRT sequencing and its implications in molecular studies of Stipa. Sci Rep. 11(1):15345.
- Baiakhmetov E, Nowak A, Gudkova PD, Nobis M. 2020. Morphological and genome-wide evidence for natural hybridisation within the genus Stipa (Poaceae). Sci Rep. 10(1):13803.
- Chase MW, Christenhusz MJM, Fay MF, Byng JW, Judd WS, Soltis DE, et al. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181(1):1–20.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Huang D, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Jia S. 1997. Flora of Chinese forage plants. Beijing: China Agriculture Press.
- Jing Z, Yu J, Cheng J. 2013. Genetic diversity of a dominant species Stipa bungeana and its conservation strategy in the Loess Plateau of China. Biochem Syst Ecol. 47:126–131.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Krawczyk K, Nobis M, Myszczyński K, Klichowska E, Sawicki J. 2018. Plastid super-barcodes as a tool for species discrimination in feather grasses (Poaceae: Stipa). Sci Rep. 8(1):1924.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kuo P. 1987. Flora of China. Vol. 9(3). Beijing: Science Press.
- Kuo P, Sun Y. 1982. A preliminary study on the classification, distribution and ecological nature of genus Stipa L. of China. J Univ Chinese Acad Sci. 20(1):34–44.
- Mishra P, Kumar A, Nagireddy A, Mani DN, Shukla AK, Tiwari R, Sundaresan V. 2016. DNA barcoding: an efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol J. 14(1):8–21.
- Myszczyński K, Nobis M, Szczecinska M, Sawicki J, Nowak A. 2016. The complete plastid genome of the middle Asian endemic of Stipa lipskyi (Poaceae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4661–4662.
- Nobis M, Gudkova PD, Baiakhmetov E, Żabicka J, Krawczyk K, Sawicki J. 2019. Hybridisation, introgression events and cryptic speciation in Stipa (Poaceae): A case study of the Stipa heptapotamica hybrid-complex. Perspect Plant Ecol Evol Syst. 39:125457.
- Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, et al. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 20(10):714–737.
- Stefanova P, Taseva M, Georgieva T, Gotcheva V, Angelov A. 2013. A modified CTAB method for DNA extraction from soybean and meat products. Biotechnol Biotec Eq. 27(3):3803–3810.
- Van Do T, Xu B, Gao X. 2021. Molecular phylogeny and character evolution of Flemingia (Leguminosae, Papilionoideae, Phaseoleae, Cajaninae) inferred from three cpDNA and nrITS sequence data. Plant Syst Evol. 307(2):30.