Abstract
Persicaria perfoliata (L.) H. Gross is an herbal medicine with a long history of common use in China. In this study, we sequenced and assembled the complete chloroplast genome sequence of P. perfoliata and investigated its phylogenetic relationship in the family Polygonaceae. The total genome size is 160,585 bp in length with 37.96% GC content, consisting of a small single-copy (SSC) of 12,876 bp, a large single-copy (LSC) of 85,439 bp, and two inverted repeats (IRs) of 31,135 bp. The cp genome contains 128 genes, including 35 tRNA genes, eight rRNA genes, and 85 protein-coding genes. The phylogenetic tree showed that P. perfoliata was closely related to P. maackiana, and Persicaria exhibited a closer relationship with Bistorta in the family Polygonaceae. This work provides a molecular basis for investigating the evolutionary status, phylogenetic relationships, and population genetics of this species.
Persicaria perfoliata (L.) H. Gross 1919 is an annual herbaceous plant belonging to the genus Persicaria of Polygonaceae (Hough-Goldstein et al. Citation2008). It is distributed in the Philippines, Indonesia, North Korea, India, Japan, Russia (Siberia), and China, exhibiting strong survival adaptability (Tan et al. Citation2013). In China, P. perfoliata has a long history being used as a traditional medicine. Modern pharmaceutical studies have shown that P. perfoliata has various effects such as antioxidation, anti-tumor, antiviral, anti-inflammatory, anti-liver fibrosis, expectorant, antitussive, and antibacterial activities (Liu et al. Citation2020). Previous studies on P. perfoliata were focused on quality control, pharmacological activities, and phytochemistry (Wang et al. Citation2009; Xing et al. Citation2011; Tian et al. Citation2013). However, up to now for such medicinal plant, the chloroplast genome of P. perfoliata has not been analyzed. In this study, we report the complete cp genome sequence of P. perfoliata and elucidate its phylogenetic position in the family Polygonaceae.
The healthy and fresh leaves of P. perfoliata were collected from Xiaoxing’an Mountains, Heilongjiang Province, China (129°39′E, 46°51′N). The specimens of P. perfoliata were deposited at the Herbarium of Jiamusi University under the voucher number XAL2021001 (contact person: Hongsheng Yang and Email: [email protected]). Total genomic DNA was extracted by the OMEGA E.Z.N.A.® Plant DNA Kit and sequenced on the Illumina NovaSeq 6000 platform with 150 bp paired-end reads. 72,376,958 raw reads were obtained. Trimmomatic (Bolger et al. Citation2014) was used to filter the low-quality reads. The remaining 72,257,936 clean reads were used to assemble the cp genome by NOVOplasty v4.3.1 (Dierckxsens et al. Citation2017) with the complete cp genome of Persicaria chinensis (L.) H. Gross (accession number: MN627221) as the reference (Yu et al. Citation2020). The program PGA (Qu et al. Citation2019) and GeSeq online (Tillich et al. Citation2017) performed the annotation followed by manual adjustments. The annotated genomic sequence was submitted to GenBank under accession number OL679838.
The complete cp genome size of P. perfoliata is 160,585 bp with 37.96% GC content, which contains two inverted repeats (IRs, 31,135 bp each), separated by small single-copy (SSC, 12,876 bp) and large single-copy (LSC, 85,439 bp) region. The corresponding GC values of the IRs, SSC, and LSC are 41.43%, 33.01%, and 36.18%, respectively. There are 128 annotated genes, including 35 transfer RNA (tRNA) genes, eight ribosomal RNA (rRNA) genes, and 85 protein-coding genes (PCGs). Nineteen genes contain one intron, and four genes (ycf3, clpP, and two rps12) contain two introns.
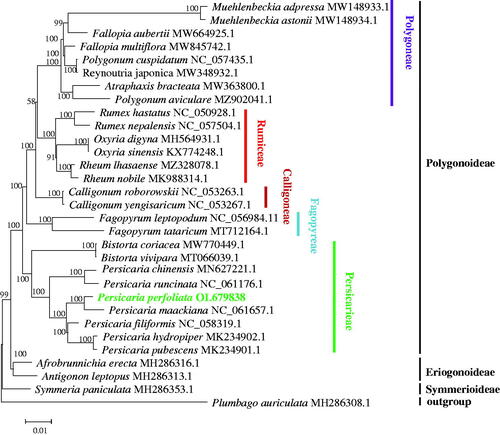
To investigate the phylogenetic status of P. perfoliata in the family Polygonaceae, a phylogenetic tree based on the complete cp genomes of P. perfoliata and 29 other species of Polygonaceae was reconstructed using Plumbago auriculata as an outgroup (). The complete cp genomes were aligned by MAFFT v7.307 (Katoh and Standley Citation2013), and using the maximum-likelihood (ML) method the phylogenetic analysis was performed by MEGA X (Kumar et al. Citation2018). The phylogenetic tree showed that P. perfoliata was closely related to P. maackiana, and Persicaria exhibited a closer relationship with Bistorta in the family Polygonaceae. Polygonoideae was divided into five tribes including Polygoneae, Rumiceae, Calligoneae, Fagopyreae, and Persicarieae based on the phylogenetic tree, which was consistent with the previous taxonomic treatment (Sanchez et al. Citation2011).
Figure 1. Phylogenetic tree of Persicaria perfoliata and 29 other species of Polygonaceae based on the complete chloroplast genome sequence. The species of Plumbago auriculata from Plumbaginaceae served as the outgroup. The tree was generated by maximum-likelihood (ML) analysis using MEGA X and the GTR + G+I nucleotide model. The stability of each tree node was tested by bootstrap analysis with 1000 replicates. The numbers on branches were bootstrap support values.
Ethics statement
The collection of Persicaria perfoliata was approved by Tuanjie forest farm, Tangyuan county, Heilongjiang province, China.
Author contributions
Lili Li and Decai Liu were involved in the conception and design. Hongsheng Yang and Donghong Yang analyzed the data and wrote the initial draft of the paper. Xuewen Yang, Qingbo Zhou, and Haitao Cheng collected the data and contributed to the revisions. All authors agreed to be accountable for all aspects of the work and approved the version to be published.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OL679838. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA786268, SRR17152982, and SAMN23638302, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hough-Goldstein J, Schiff M, Lake E, Butterworth B. 2008. Impact of the biological control agent Rhinoncomimus latipes (Coleoptera: Curculionidae) on mile-a-minute weed, Persicaria perfoliata, in field cages. Biol Control. 46(3):417–423.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu JY, Zeng YL, Sun GJ, Yu SP, Xu Y, He C, Li ZL, Jin SR, Qin XH. 2020. Polygonum perfoliatum L., an excellent herbal medicine widely used in China: a review. Front Pharmacol. 11:581266.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Sanchez A, Schuster TM, Burke JM, Kron KA. 2011. Taxonomy of Polygonoideae (Polygonaceae): a new tribal classification. Taxon. 60(1):151–160.
- Tan JC, Sun QW, Wei SH, Ran MX, Dang HJ, Yang YL. 2013. Wild resources and suitable plangting area of Polygonum perfoliatum. Guizhou Agric Sci. 41(2):16–19.
- Tian L, Zhao Y, Zhou X, Chen HG, Zhao C, Gong XJ. 2013. Studies on chromatographic fingerprint and fingerprinting profile–efficacy relationship of Polygoni Perfoliati Herba. Evid Based Complement Altern Med. 2013:1–15.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq– versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang Q, Chen L, Tian Y, Li B, Sun QH, Dong JX. 2009. Chemical constituents of Polygonum perfoliatum L. Bull Acad Milit Med Sci. 33:254–256.
- Xing YJ, Wang HY, Wang JX, Li YY, Kang WY. 2011. α-Glucosidase inhibitory and antioxidant activities of Polygonum perfoliatum. Chin J Exp Tradit Med Formul. 17(2):189–191.
- Yu BX, Liu J, Liu X, Lan XZ, Qu XY. 2020. The complete chloroplast genome sequence of Polygonum chinense L. Mitochondrial DNA B Resour. 5(3):2139–2140.
