Abstract
A species of Cardamine circaeoides Hook. f. et Thoms., 1861, which belongs to the Cardamine family (Brassicaceae), is an endemic species of the Wuling Mountains in Hunan and Hubei Provinces of China. Since there are many morphologically related species of C. circaeoides, the chloroplast (cp) genome characteristics of C. circaeoides were analyzed in order to explore the phylogenetic relationship between it and the closely related species. A cp genome totaling 154,838 base pairs exhibited a typical quadripartite structure with a pair of IRs (inverted repeats; 26,493 base pairs) separated by a small single-copy region of 17,938 base pairs and a large single-copy region of 83,914 base pairs. A total of 130 genes were found in the cp genome, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. There were 36.21%, 33.96%, 29.09%, and 42.35% GC content in the entire cp genome, LSC region, SSC region, and IR region, respectively. According to phylogenetic analysis, C. circaeoides is evolutionarily closer to Cardamine hupingshanensis.
Cardamine circaeoides belongs to the Brassicaceae family. The species is primarily found in the high mountains, valleys, and streams of the Wuling mountains of Hunan and Hubei Provinces, China (Wu et al. Citation2020). Besides being rich in protein, soluble sugar, amino acids, cellulose, and other nutrients, it also provides a super enrichment of selenium (Tian and Cheng Citation2003). In the case of selenium supplement cultivation, organic selenium concentrations can reach 3553 mg/kgDW (Both et al. Citation2018) and its tender stems and leaves can be eaten. In the field survey, local residents often refer to it as ‘wild rape’, ‘water rape’, or ‘rock plate vegetable’, which is a highly nutritious wild vegetable. As technology continues to develop rapidly, scientists began to pay attention to the application of modern molecular techniques in order to characterize the genetic diversity of shepherd’s purses. It has been demonstrated in several research studies that chloroplast (cp) genome sequences play an important role in plant phylogeny and genetic analysis (Wang et al. Citation2016; Ma et al. Citation2021; Yu et al. Citation2021). Cardamine circaeoides’s cp genome has been reported for the first time, and its structural characteristics have been analyzed. This study provides important genetic and phylogenetic information for understanding this species’ genetic relationships.
This study examined the fresh leaves of C. circaeoides collected from Enshi City, Hubei Province, China (30.301 N, 109.487E, Altitude:1236 m). A specimen was deposited at the Guangdong Agro-biological Gene Research Center (http://multi-omics.agrogene.ac.cn/, contact person:Yongjian Luo, and email: [email protected]) under the voucher number 20210303005. An extraction of genomic DNA from dried silica leaves was performed using Qiagen’s DNeasy Plant Mini Kit. The 1% agarose gel and Thermo Scientific Nanodrop spectrophotometer were used to confirm the quality and quantity of the samples (). A high-throughput sequencing platform (HiSeq 4000) was used to sequence the complete genomes of C. circaeoides. The paired-end sequence reads were trimmed to remove adaptors and low-quality sequences via Trimmomatic 0.39. Clean reads were assembled using GetOrganelle v1.5 (Jin et al. Citation2020). Chloroplast genome annotation was performed using CPGAVAS2 (http://47.96.249.172:16019/analyzer/home) (Shi et al. Citation2019) and GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017). An accession number (OL634846) has been assigned to the annotated cp genomic sequence in GenBank.
Figure 1. Morphological characteristics of wild conditions in C. circaeoides. Species photos were taken by the author in Wuyi Mountain, Enshi, Hubei, China, June 2020, without any copyright issues.

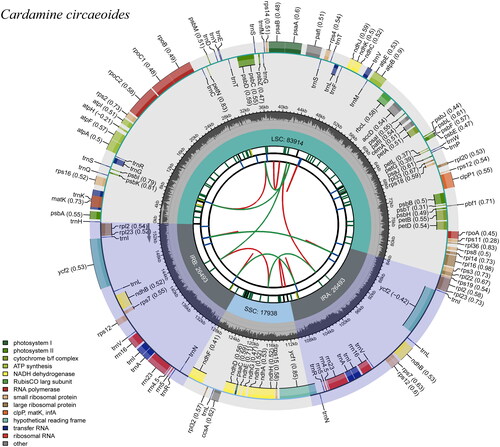
There are 154,838 base pairs in the cp genome of C. circaeoides (). In the circular cp genome of C. circaeoides, a quadripartite structure was observed with an LSC (83,914 bp), an SSC (17,938 bp), and two IR regions (IRA and IRB, each of 26,493 bp). A total of 36.21% of C. circaeoides’ cp genome is composed of GCs (LSC, 33.96%; SSC, 29.09%; IR, 42.35%). GC content and gene order were similar to those of Cardamine amaraeform (Raman et al. Citation2021). A total of 130 genes were found in the cp genome, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The introns were present in the following 12 protein-coding genes (atpF, clpP1, ndhA, ndhB, pafI, petB, petD, rpl16, rpl2, rpoC1, rps16), and 6 tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC). In C. circaeoides, 13 cis-clipping genes including rps16, atpF, rpoC1, pafI, clpP1, petB, petD, rpl16, rpl2(2), ndhB(2), ndhA, and a trans-splicing gene rps12 were detected by CPGview-RSG (http://www.herbalgenomics.org/cpgview/). In addition, 95 SSRs were identified, including 89 single nucleotides (A/C/T, 93.68%) and 5 double nucleotides (AT/TA, 6.32%). At the same time, 23 tandem repeats were identified, with a total length of 15–42 bp. 33 dispersed repeats were identified with a threshold of 1E-4, and the length of repeat units ranged from 30 to 67.
Figure 2. Graphic representation of features identified in C. circaeoides plastome by using CPGview-RSG (http://www.herbalgenomics.org/cpgview). The map contains seven circles. From the center going outward, the first circle shows the distributed repeats connected with red (the forward direction) and green (the reverse direction) arcs. The next circle shows the tandem repeats marked with short bars. The third circle shows the microsatellite sequences as short bars. The fourth circle shows the size of the LSC and SSC. The fifth circle shows the IRA and IRB. The sixth circle shows the GC contents along the plastome. The seventh circle shows the genes having different colors based on their functional groups.
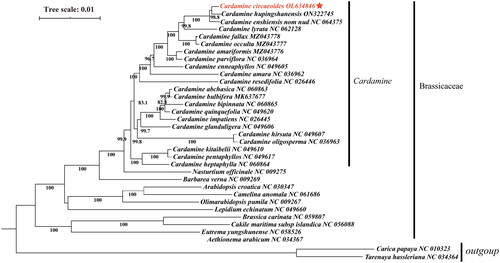
For the purpose of analyzing the phylogenetic placement of C. circaeoides within Cardamine, we used the cp genome sequence of C. circaeoides and 31 other related Brassicaceae species, Carica papaya, and Tarenaya hassleriana interior as an outgroup to construct the phylogenetic tree. In order to align the sequences, MAFFT v7.450 was used with the default parameters (Katoh and Standley Citation2013). A maximum likelihood (ML) tree was then constructed using MEGE version X with a Generalized Time-Reversible (GTR) model, with 1000 bootstrap replications. According to the phylogenetic tree (), the species of the Cardamine genera are monophyletic. Phylogenetic analysis demonstrated that C. circaeoides is closely related to Cardamine hupingshanensis with a strong bootstrap value (98.8% for ML).
Ethical approval
Neither the samples used in this study nor the sampling site are listed on either the IUCN Red List of Threatened Species or the List of State-protected Plant Species. Both the field study and laboratory study were conducted in accordance with the guidelines established by Hubei Minzu University.
Authors’ contributions
Yongjian Luo and Zhijun Deng designed the experiment and approved the final version of the paper. Zhijun Deng and Ru Wang wrote and revised the paper; Zhijun Deng Yongjian Luo and Ru Wang analyzed and interpreted the data. All authors are responsible for all aspects of their work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
These findings are supported by genome sequence data available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under accession number OL634846, associated BioProject, SRA, Bio-Sample numbers are PRJNA867123, SRR20954704, and SAMN30184410, respectively.
Additional information
Funding
References
- Both EB, Shao S, Xiang J, Jókai Z, Yin H, Liu Y, Magyar A, Dernovics M. 2018. Selenolanthionine is the major water-soluble selenium compound in the selenium tolerant plant C. circaeoides. Biochim Biophys Acta Gen Subj. 1862(11):2354–2362.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Ma C-X, Yan H-F, Ge X-J. 2021. The complete chloroplast genome of Lithocarpus hancei (Benth.) Rehd (Fagaceae) from Zhejiang, China. Mitochondrial DNA B Resour. 6(7):2022–2023.
- Raman G, Park KT, Park S. 2021. The complete chloroplast genome of an endemic plant to Korea, Cardamine amaraeformis Nakai.: genome structure and phylogenetic analysis. Mitochondrial DNA B Resour. 6(9):2725–2726.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Tian L, Cheng L. 2003. Analysis of nutrients in Cardamine hupingxhanensis and its cultivation in foreign land. Sheng Ming ke Xue Yan Jiu. 7(2):167–172.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang L, Wuyun T, Du H, Wang D, Cao D. 2016. Complete chloroplast genome sequences of Eucommia ulmoides: genome structure and evolution. Tree Genet Gen. 12(1):12.
- Wu M, Cong X, Li M, Rao S, Liu Y, Guo J, Zhu S, Chen S, Xu F, Cheng S, et al. 2020. Effects of different exogenous selenium on Se accumulation, nutrition quality, elements uptake, and antioxidant response in the hyperaccumulation plant. Ecotoxicol Environ Saf. 204:111045.
- Yu Y, Ouyang Z, Guo J, Zeng W, Zhao Y, Huang L. 2021. Complete chloroplast genome sequence of Erigeron breviscapus and characterization of chloroplast regulatory elements. Front Plant Sci. 12:758290.