Abstract
The little-known genus Pseudoechthistatus Pic, 1917 belongs to the subfamily Lamiinae of the family Cerambycidae. Adult Pseudoechthistatus hei Xie and W. Wang, 2019 specimens were collected from Bāijì Hill, Xīntángfáng Village, Wéixī County, Yúnnán Province, China. The complete mitochondrial genome (GenBank accession number: ON641973.1) of P. hei was sequenced, annotated, and characterized; it is a circular DNA molecule of 16,103 bp with a 75.71% AT content, and it comprised 13 protein-coding genes (PCG), 22 tRNA genes, two rRNA genes, and 1 control region. The PCGs initiated with the typical ATN (Met) start codons, and were terminated by typical TAN stop codons. The Bayesian Inference phylogenetic tree was first constructed using JTT + F + I + G4 model for P. hei, which showed that P. hei was closely related to Monochamus alternatus alternatus.
Introduction
The little-known genus Pseudoechthistatus Pic, 1917 belongs to the tribe Lamiini of the family Cerambycidae (Bi and Lin Citation2016), which is unique because of its conspicuously raised sub-basal tubercle on each elytron (Bi and Lin Citation2016). Pseudoechthistatus hei Xie and W. Wang, 2019 has a significantly reduced preapical stripe on the elytron (nearly presenting a very short and narrow longitudinal spot) (Wang P et al. Citation2019; ). Currently, the nucleotide database of the National Center for Biotechnology Information (NCBI) contains publicly available information on the mitochondrial genome of the genus Pseudoechthistatus. In this study, we first have sequenced and annotated the complete mitochondrial genome (mitogenome) of P. hei and constructed phylogenetic trees, which can contribute to the understanding of its mitogenome characteristics and determination of the phylogenetic position of the genus Pseudoechthistatus.
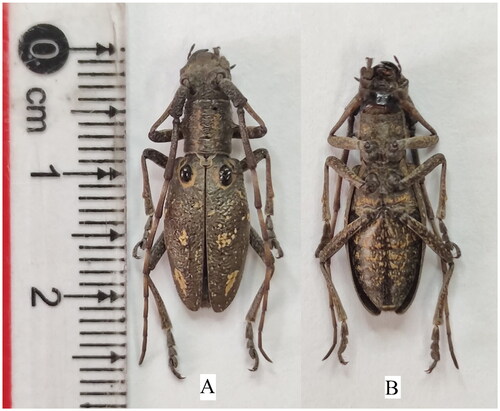
Figure 1. Reference images of adult female of Pseudoechthistatus hei. (A) the ventral view of P. hei; (B) the dorsal view of P. hei. These photographs from our specimens were taken by us. The sex mark including the antennae was about 2.1 times as long as the body of the male while it was 1.3 times as long as the body of the female (Wang P et al. Citation2019).
Materials
Adult P. hei specimens were collected from Bāijì Hill (99.22° N, 27.42° E), Xīntángfáng Village, Wéixī County, Yúnnán Province, China, on May 21, 2021, and deposited in the animal specimen room of Guiyang University (http://en.gyu.cn/, Yu Bai, [email protected]) under the voucher number GYU-20210521-001.
Methods
Genomic DNA was isolated using the Qiagen DNeasy Blood and Tissue Extraction kit (Qiagen, Germantown, MD, USA) and subjected to paired-end sequencing (2 × 150 bp) of 300 bp inserts using an Illumina NovaSeq 6000 platform (Illumina, Inc., San Diego, CA, USA). The raw reads were filtered using fastp v0.23.2 (https://github.com/OpenGene/fastp) (Chen et al. Citation2018). Quality control (QC) standards of reads from DNA were as follows: (1) trimming adapter sequences with >6 bases, (2) removing reads with >0 unidentified nucleotides (N), (3) removing reads with >20% bases with Phred quality < Q30, and (4) removing reads with <150 bases. The genome was assembled de novo using NOVOPlasty v4.3.1 (https://github.com/ndierckx/NOVOPlasty) (Dierckxsens et al. Citation2017) with default parameters and the mitogenome of Monochamus sparsutus (GenBank accession number: MW067124) as a seed sequence. The AT-skew [(A − T)/(A + T)] and GC-skew [(G − C)/(G + C)] of the sequence were estimated to investigate the nucleotide composition bias using Perna and Kocher’s formula (Perna and Kocher Citation1995). The P. hei mitogenome was initially annotated using GeSeq Version 2.03 (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017), using the third-party software tRNAscan-SE v2.0.7 (Chan and Lowe Citation2019), ARWEN v1.2.3 (Laslett and Canbäck Citation2008), and BLAT v36x7 (Kent Citation2002) with the mitogenome of Monochamus sparsutus as a reference. The start and stop codons of the protein-coding genes (PCG) were corrected manually using the mitogenomes of M. sparsutus, M. alternatus alternatus (MT547196) (Liao et al. Citation2020) and M. alternatus (KJ809086) (Li F et al. Citation2016) as references. The order and orientation of the genes were determined and plotted using the OGDRAW web server (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Greiner et al. Citation2019). For phylogenetic analyses, the mitogenomes of 19 Lamiinae species and two outgroup species were used to construct a Bayesian Inference (BI) phylogenetic tree using PhyloSuite version 1.2.2 (Zhang D et al. Citation2020) with MAFFT version 7 (Katoh and Standley Citation2013) and MrBayes version 3.2.7 (Ronquist et al. Citation2012). Reference sequences of the subfamily Lamiinae published in formal publications were selected from the NCBI nucleotide database. The amino acid sequences of 13 PCGs in their mitogenomes were aligned using MAFFT with default parameters. According to Bayesian Information Criterion (BIC) scores, JTT (Jones-Taylor-Thornton) + F [State frequencies, dirichler (1.0,1.0,1.0,1.0)] + I [Proportion of invariable sites, uniformly distributed on the interval (0.00,1.00)] + G4 (gamma-distributed rate variation, four categories) model was selected as the best-fit partition model (edge-unlinked) using ModelFinder (Kalyaanamoorthy et al. Citation2017) for BI of amino acid sequences. In the BI analysis, two runs of 455,000 generations were conducted for each matrix, and the initial 25% were discarded as burn-in, which had the a same topology with an average standard deviation of split frequencies was 0.008362 (<0.01). The resulting phylogenetic tree is shown using FigTree 1.4.4 (https://github.com/rambaut/figtree/).
Results
We obtained approximately 43.88 Gb (97.76%) of clean high-quality data from 51.34 Gb of raw data. The circular mitogenome (GenBank accession number: ON641973.1) of P. hei was completely assembled and was 16,103 bp (nucleotide composition: 38.74% A, 36.96% T, 9.45% G, and 14.85% C) with 75.71% AT content. The AT- and GC-skew of the major strand of the mitogenome were estimated to be 0.0235 and −0.2224, respectively. The mitogenome of P. hei was comprised of 13 PCGs, 1 control region (CR), 22 tRNA genes, and two rRNA genes ().
Figure 2. Mitogenome pattern map of Pseudoechthistatus hei. Grey arrows indicate the direction of gene transcription (5'→3'). Genes inside the black circle are coded in the majority strand (N-strand); genes outside the black circle are coded in the minority strand (J-strand).
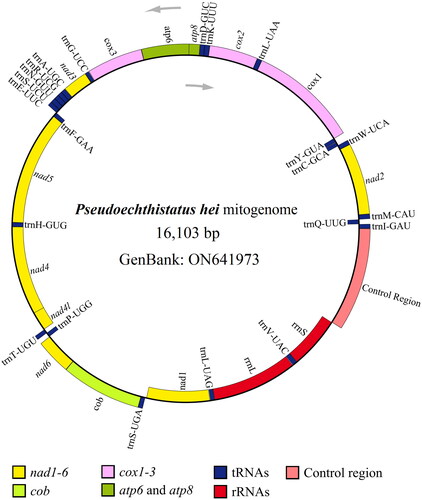
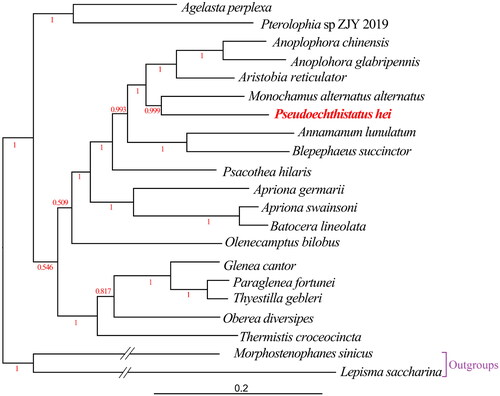
All 13 PCGs had a typical ATN (Met) start codon: two PCGs (nad5 and nad1) initiated with an ATA start codon, five PCGs (cox1, cox2, atp8, nad3, and nad6) initiated with an ATT start codon, five PCGs (atp6, cox3, nad4, nad4l, and cob) initiated with an ATG start codon, and only one PCG (nad2) initiated with an ATC start codon. All 13 PCGs contained a typical TAN stop codon: two PCGs (cob and nad1) terminated with a TAG stop codon, seven PCGs (nad2, atp8, atp6, nad3, nad5, nad4l, and nad6) ended with a TAA stop codon, and four PCGs (cox1, cox2, cox3, and nad4) terminated with an incomplete stop codon (T), consisting of a codon that was completed by the addition of A nucleotides at the 3′ end of the encoded mRNA. The 22 tRNA ranged from 56 bp (trnC-GCA) to 72 bp (trnK-CUU) in length. The rrnL and rrnS genes were 1,276 and 785 bp in length, respectively. The CR, also an AT-rich region, was 1,460 bp in length, with an AT content of 80.55%, and was located between the rrnS and trnI-GAU genes. Based on the amino acid sequences of 13 PCGs from 21 mitogenomes, a phylogenetic tree was constructed using BI method (), the structure of which was similar to that reported in previous studies (Liao et al. Citation2020). The results showed that P. hei was closely related to Monochamus alternatus alternatus with high support value (posterior probability value = 0.999).
Figure 3. Bayesian Inference (BI) phylogenetic tree from amino acid sequences of 13 PCGs of 21 mitogenomes using MrBayes under the JTT + F + I + G4 model. BI posterior probability values were shown in red color. The complete mitogenome of Pseudoechthistatus hei (ON641973) determined in this study is indicated in red color. The branches of outgroups are depicted as half of their original branch length. The following sequences were used: Apriona germari MW858151 (Zhang Z-Y et al. Citation2021), Paraglenea fortunei MW858148 (Zhang Z-Y et al. Citation2021), Annamanum lunulatum MN356095 (Dai et al. Citation2020), Aristobia reticulator MK423971 (Behere et al. Citation2019), Anoplophora chinensis KT726932 (Li W et al. Citation2016), Oberea diversipes MN709785 (Tian and Wang Citation2020), Agelasta perplexa MW067123 (Li et al. Citation2021), Olenecamptus bilobus MT740324 (Dong et al. Citation2021), Monochamus alternatus alternatus MT547196 (Liao et al. Citation2020), Batocera lineolata JN986793 (Wang C et al. Citation2012), Psacothea hilaris FJ424074 (Ki-Gyoung et al. Citation2009), Glenea cantor MN044086 (Wang X et al. Citation2019), Apriona swainsoni KX184801 (Que et al. Citation2019), Anoplophora glabripennis DQ768215 (Fang et al. Citation2016), Thyestilla gebleri KY292221 (Yang et al. Citation2017), Pterolophia sp. ZJY-2019 MK863510 (Wang J et al. Citation2019), Thermistis croceocincta MK863511 (Wang J et al. Citation2019), Blepephaeus succinctor MK863507 (Wang J et al. Citation2019), Morphostenophanes sinicus MW853764 (Bai et al. Citation2021) and Lepisma saccharina MT108230 (Bai et al. Citation2020).
Discussion and conclusion
Although PCGs of the P. hei mitogenome only had typical ATN (Met) start codons, TTG, GTA and ATN were used as the start codons of nad1 and nad3 in mitogenomes of subfamily Lamiinae (Wang C-Y et al. Citation2013; Li F et al. Citation2016; Zhang Z-Y et al. Citation2021), which reflects the evolutionary diversity of the species and makes it difficult to determine the start and stop positions of PCGs. Taken together, the complete mitogenome of P. hei can contribute to the understanding of the mitogenome characteristics and determination of the phylogenetic position of the genus Pseudoechthistatus.
Ethics statement
This research does not involve ethical research. Insects are invertebrates, and there are no ethics involved in using them in experiments.
Authors’ contributions
Yu Bai, analyzed the data, uploaded the analysis data, involved in certain tools for analysis, drafted of the paper, and approved the final draft.
Lin Ye, collected and analyzed data.
Kang Yang, performed the experiments and analyzed the data.
Hui Wang, identified insects, contributed reagents/materials, involved in conception and design of the work, performed the experiments, prepared figure, and approved and published the final draft.
All authors agree to be accountable for all aspects of the work.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. ON641973.1. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA857171, SAMN29620688, and SRR20067649, respectively.
Additional information
Funding
References
- Bai Y, Chen J, Li G, Wang H, Luo J, Li C. 2020. Complete mitochondrial genome of the common silverfish Lepisma saccharina (Insecta: Zygentoma: Lepismatidae). Mitochondrial DNA Part B. 5(2):1552–1553.
- Bai Y, Gao X, Yu Y, Long X, Zeng X, Wei D, Ye L. 2021. Complete mitochondrial genome of Morphostenophanes sinicus (Zhou, 2020) (Insecta: Coleoptera: Tenebrionidae). Mitochondrial DNA B Resour. 6(10):2946–2948.
- Behere GT, Tay WT, Firake DM, Kunz D, Burange PS, Ramamurthy VV. 2019. Characterization of draft mitochondrial genome of guava trunk borer, Aristobia reticulator (Fabricius, 1781) (Coleoptera: Cerambycidae: Lamiinae) from India. Mitochondrial DNA Part B. 4(1):1592–1593.
- Bi W-X, Lin M-Y. 2016. A revision of the genus Pseudoechthistatus Pic (Coleoptera, Cerambycidae, Lamiinae, Lamiini). ZK. 604:49–85.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. In: Kollmar M, editor. Gene prediction: methods and protocols. New York (NY): Springer New York; p. 1–14.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Dai X-Y, Zhang H, Xu X-D, Jia Y-Y, Zhang J-Y, Yu D-N, Cheng H-Y. 2020. The complete mitochondrial genome of Annamanum lunulatum (Coleoptera: lamiinae) and its phylogeny. Mitochondrial DNA B Resour. 5(1):551–553.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18-e18.
- Dong Z, Wang X, Li Y, Huang D, Lu W. 2021. Complete mitochondrial genome of an Asian longicorn beetle, Olenecamptus bilobus Fabricius (Coleoptera: cerambycidae: lamiinae). Mitochondrial DNA B Resour. 6(2):560–561.
- Fang J, Qian L, Xu M, Yang X, Wang B, An Y. 2016. The complete nucleotide sequence of the mitochondrial genome of the Asian longhorn beetle, Anoplophora glabripennis (Coleoptera: cerambycidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3299–3300.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12(4):656–664.
- Ki-Gyoung K, Mee Yeon H, Min Jee K, Hyun Hwak I, Man Il K, Chang Hwan B, Sook Jae S, Sang Hyun L, Iksoo K. 2009. Complete mitochondrial genome sequence of the yellow-spotted long-horned beetle Psacothea hilaris (Coleoptera: Cerambycidae) and phylogenetic analysis among coleopteran insects. Mol Cells. 27(4):429–441.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Li Y, Chen R, Jia H, Lei X, Ma J, Wu S. 2021. The complete mitochondrial genome of Agelasta perplexa Pascoe (Coleoptera: cerambycidae). Mitochondrial DNA B Resour. 6(6):1694–1695.
- Li W, Yang X, Qian L, An Y, Fang J. 2016. The complete mitochondrial genome of the citrus long-horned beetle, Anoplophora chinensis (Coleoptera: cerambycidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4665–4667.
- Li F, Zhang H, Wang W, Weng H, Meng Z. 2016. Complete mitochondrial genome of the Japanese pine sawyer, Monochamus alternatus (Coleoptera: cerambycidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1144–1145.
- Liao Q, Yang X, Ma J, Sheng L, Zou S. 2020. The complete mitochondrial genome of Monochamus alternatus alternatus (Coleoptera: cerambycidae). Mitochondrial DNA B Resour. 5(3):3381–3382.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41(3):353–358.
- Que S, Yu A, Liu P, Jin M, Xie G. 2019. The complete mitochondrial genome of Apriona swainsoni. Mitochondrial DNA Part B. 4(1):931–932.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Tian L, Wang J. 2020. Complete mitochondrial genome of Oberea diversipes (Coleoptera, Cerambycidae). Mitochondrial DNA B Resour. 5(1):375–376.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang J, Dai X-Y, Xu X-D, Zhang Z-Y, Yu D-N, Storey KB, Zhang J-Y. 2019. The complete mitochondrial genomes of five longicorn beetles (Coleoptera: cerambycidae) and phylogenetic relationships within Cerambycidae. PeerJ. 7:e7633.
- Wang C, Feng Y, Chen X. 2012. Complete sequence and gene organization of the mitochondrial genome of Batocera lineolata Chevrolat (Coleoptera: cerambycidae). Chin Sci Bull. 57(27):3578–3585.
- Wang C-Y, Feng Y, Chen X-M. 2013. Complete coding region of the mitochondrial genome of Monochamus alternatus Hope (Coleoptera: cerambycidae). Zoolog Sci. 30(7):570–576. 577.
- Wang P, Xie G, Wang W. 2019. A new species of Pseudoechthistatus Pic, 1917 (Coleoptera, Cerambycidae, Lamiinae) from Yunnan, China. Zootaxa. 4619(1):zootaxa.4619.1.10–4610. zootaxa4619eng.
- Wang X, Zheng X, Lu W. 2019. The complete mitochondrial genome of an Asian longicorn beetle Glenea cantor (Coleoptera: Cerambycidae: Lamiinae). Mitochondrial DNA B Resour. 4(2):2906–2907.
- Yang J, Cao L, Geng Y, Wei D, Chen M. 2017. Determination of the complete mitochondrial genome of Thyestilla gebleri and comparative analysis of the mitochondrial genome in Cerambycidae. Chinese Journal of Applied Entomology. 54(5):755–766.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhang Z-Y, Guan J-Y, Cao Y-R, Dai X-Y, Storey KB, Yu D-N, Zhang J-Y. 2021. Mitogenome analysis of four Lamiinae species (Coleoptera: Cerambycidae) and gene expression responses by Monochamus alternatus when infected with the parasitic nematode, Bursaphelenchus mucronatus. Insects. 12(5):453.
