Abstract
Clematis chinensis Osbeck is an important medicinal and edible plant. The complete chloroplast genome of C. chinensis Osbeck was constructed and annotated for the first time in this study. Full length of the chloroplast genome of C. chinensis Osbeck is 159,647 bp, with a large single-copy (LSC) region of 86,301 bp, a small single-copy (SSC) region of 79,536 bp, and a pair of inverted repeats IRa and IRb regions of 31,039 bp. The result of the gene annotation identified the 135 genes in the chloroplast genome, including 91 protein-coding genes, 36 tRNA genes, and eight rRNA genes. The total amount of GC is 47.82%. In the phylogenetic analysis, C. chinensis Osbeck showed the closest relationship with Clematis uncinata.
Clematis chinensis Osbeck 1757 is an important medicinal plant which is one of the origin plants of Clematidis Radix et Rhizoma (‘Weilingxian’ in Chinese) (Jiang, Pan, et al. Citation2022). The roots of this plant are used for anti-inflammatory analgesic and anti-tumor as a traditional Chinese medicine (Ye et al. Citation2021). In Yao medicine (YM) of China, the root of C. chinensis Osbeck is also called ‘Hei-jiu-niu’. It has been used widely to treat various diseases/disorders, such as relieving rheumatism pain and scapulohumeral periarthritis, etc. (Koo et al. Citation2020; Jiang, Guo, et al. Citation2022). Crude extracts and isolated compounds of C. chinensis Osbeck have been reported to have a wide range of pharmacological activities (Lin et al. Citation2021). Toxicological studies have shown that C. chinensis Osbeck can cause oral burning, abdominal pain or severe diarrhea, swelling, difficulty breathing, and dilated pupils (Lin et al. Citation2021). According to the YM theory, this herb could dispel and remove the ‘evil wind and dampness’ of arthritic patients. As an essential herb for promoting the biological function of chondrocytes (Pan et al. Citation2019), C. chinensis Osbeck has been used as a component in the equation, or as an agent of arthritis treatment in China for a long time. But genetic information of C. chinensis Osbeck is still lacking. In this study, Illumina technology was applied to the sequence, the whole chloroplast genome of C. chinensis Osbeck was assembled and annotated.
Five pieces of fresh C. chinensis Osbeck leaves were collected from Jinxiu Yao Autonomous County, Guangxi Province, China (N: 23°56′6″, E: 110°14′30). A specimen was stored at the School of Food and Biochemical Engineering of Guangxi Science & Technology Normal University (https://www.gxstnu.edu.cn/, the contact person is Song Guo and the email is [email protected]) under the voucher number HJN202006. Genomic DNA from five pieces of young leaves was extracted by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. Six microgram of DNA was used as a template to construct an array library using the Illumina Hiseq 2000 platform (Illumina, San Diego, CA). Low quality reads and adapters were removed in FastQC software (Andrews Citation2010).
The chloroplast genome was assembled using the program NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017), with the complete chloroplast genome of C. aethusifolia chloroplast genome (GenBank accession number: MK253462) as the reference. After combining the paired end clean reads, a total of 17,387,048 sequences in quantity were obtained for the chloroplast genome assembly. The annotation was mainly carried out by comparing the chloroplast genomes of species with close phylogenetic relationships by Geneious v 11.1.5 (Biomatters Ltd, Auckland, New Zealand), and the results of annotation were confirmed and modified by the CPGAVAS online tool (Zuo et al. Citation2017; Wang et al. Citation2018). The annotated genomic sequence was registered in the GenBank with an accession number (MW829280). The plastome of C. chinensis Osbeck, which is a circular DNA molecule, is 159,647 bp in length. Whole chloroplast genome consisted of a large single-copy (LSC, 79,536 bp) region, a small single-copy (SSC, 18,033 bp) region, and two inverted repeat regions (Ira and Irb, 31,039 bp). Overall GC content of C. chinensis Osbeck chloroplast genome is 37.9%, with corresponding GC values of LSC, SSC, and IR regions for 36.2%, 31.3%, and 42.0%, respectively. In total, 135 unique genes were identified from the chloroplast genome of C. chinensis Osbeck, in which 91 protein-coding genes, 36 tRNA genes, and eight rRNA genes were included.
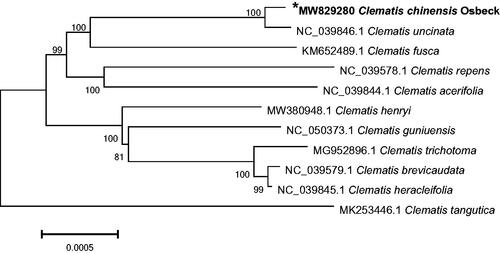
Phylogenetic analysis was performed using chloroplast genome sequences of C. chinensis Osbeck and other 10 species within the Clematis genus of Ranunculaceae family (). Phylogenetic tree was generated based on the whole chloroplast genome sequences (Zhou et al. Citation2019). Phylogenetic tree was analyzed with MEGA6 software (Koichiro et al. Citation2013) using maximum-likelihood (ML) method (bootstrap values were calculated out of 1000 replicates) (Hu et al. Citation2019). In the phylogenetic analysis, C. chinensis Osbeck was clustered closely related to Clematis uncinata (GenBank accession number: NC 039846.1) accompanied by strong bootstrap support for the Clematis section of genus Clematis family of Ranunculaceae. Our data may be useful for further studies of Clematis phylogeny and the molecular identification of promising medicinal plant.
Ethical approval
Ethical approval for the study was obtained from the Ethical Committee of Guangxi Science and Technology Normal University. The collection of specimen conformed to the requirement of International Ethics, which did not cause damage to the local environment. The process and purpose of this experimental research were in line with the rules and regulations of our institute. There are no ethical issues and other conflicts of interest in this study.
Author contributions
Song Guo and Wuwei Wu involved in the conception, design, and financial support; Zeyang Li and Mingxian He collected the sample; Yu Liu, Zeyang Li, and Song Guo analyzed the data. Wuwei Wu was involved in the drafting of the paper; Song Guo revised the manuscript and final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number MW829280. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA715070, SRR13985459, and SAMN18325372, respectively.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Cambridge (UK): Babraham Bioinformatics, Babraham Institute.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hu H-L, Zhang J-Y, Li Y-P, Xie L, Chen D-B, Li Q, Liu Y-Q, Hui S-R, Qin L. 2019. The complete chloroplast genome of the daimyo oak, Quercus dentata Thunb. Conserv Genet Resour. 11(4):409–411.
- Jiang S-Q, Guo Z-J, Pan T, Xu X-X, Yang Y-N, Wang H-Y, Li P, Li F. 2022. The multi-omics and analysis to reveal thermal processing enhanced anti-rheumatoid arthritis efficacy of Radix Clematidis in rats. J Pharm Biomed Anal. 215:114760.
- Jiang SQ, Pan T, Yu JL, Zhang Y, Wang T, Li P, Li F. 2022. Thermal and wine processing enhanced Clematidis Radix et Rhizoma ameliorate collagen II induced rheumatoid arthritis in rats. J Ethnopharmacol. 288:114993.
- Koichiro T, Glen S, Alan F, Sudhir K. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30(12):5.
- Koo B, Bae HJ, Goo N, Kim J, Kim J, Cai M, Jung IH, Cho K, Jung SY, Chang SW. 2020. A botanical drug composed of three herbal materials attenuates the sensorimotor gating deficit and cognitive impairment induced by MK-801 in mice. J Pharm Pharmacol. 72(1):149–160.
- Lin T-F, Wang L, Zhang Y, Zhang J-H, Zhou D-Y, Fang F, Liu L, Liu B, Jiang Y-Y. 2021. Uses, chemical compositions, pharmacological activities and toxicology of Clematidis Radix et Rhizome – a review. J Ethnopharmacol. 270:113831.
- Pan Y-l, Ma Y, Guo Y, Tu J, Guo G-p, Ma SM, Zheng S-y, Tu P-c, Yu B-b, Huang G-c. 2019. Effects of Clematis chinensis Osbeck mediated by low-intensity pulsed ultrasound on transforming growth factor-β/Smad signaling in rabbit articular chondrocytes. J Med Ultrasonics. 46(2):177–186. DOI: 10.1007/s10396-018-0920-z.
- Wang Q, Jiao L, Wang S, Chen P, Bi L, Zhou D, Yao J, Li J, Chen Z, Jia Y. 2018. Maintenance chemotherapy with Chinese herb medicine formulas vs. with placebo in patients with advanced non-small cell lung cancer after first-line chemotherapy: a multicenter, randomized, double-blind trial. Front Pharmacol. 9:1233.
- Ye S, Yan L, Sun YP, Li XM, Si-Yi W, Pan J, Guan W, Liu Y, Naseem A, Yang B-Y, et al. 2021. Two new terpenes from the aerial parts of Clematis chinensis Osbeck. Nat Prod Res. 36:1–8.
- Zhou Q, Chen Y, Dai J, Wang F, Wu W, Fan Q, Zhou R, Ng WL. 2019. The chloroplast genome of Chunia bucklandioides (Hamamelidaceae): a rare tree endemic to Hainan, China. Conserv Genet Resour. 11(4):427–429.
- Zuo L-H, Shang A-Q, Zhang S, Yu X-Y, Ren Y-C, Yang M-S, Wang J-M. 2017. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: genome comparative and taxonomic position analysis. PLOS One. 12(2):e0171264.