Abstract
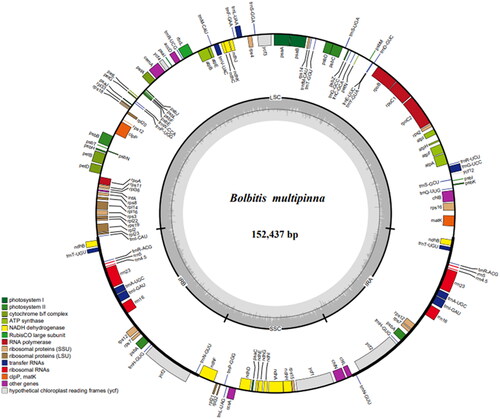
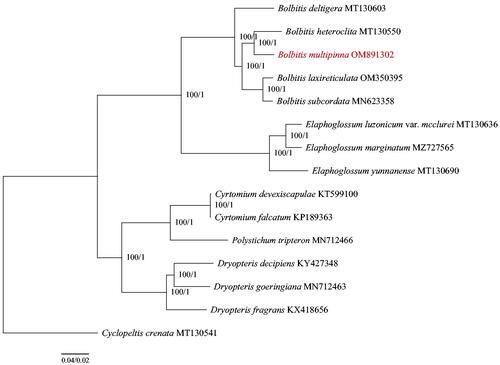
Bolbitis multipinna is an endemic fern found in Yunnan, China. In the case of the chloroplast genome, Illumina Hiseq X-Ten was used, and it revealed a conserved quadripartite structure of 152,437 bp, including a large single-copy (LSC) region of 82,618 bp, a small single-copy (SSC) region of 21,403 bp, and a pair of inverted repeat regions of 24,208 bp (IRa and IRb). The chloroplast genome of B. multipinna contains 132 genes, including 89 protein-coding genes, 35 tRNA genes, and eight rRNA genes. By using the maximum-likelihood method and the Bayesian analysis on the chloroplast genomes of 15 species ferns, the phylogenetic analysis revealed that B. multipinna is closely related to B. heteroclita.
With about 80 species, Bolbitis is a pan-tropical genus mainly found in Asia and Pacific islands (Moran et al. Citation2010; Zhang et al. Citation2013; PPG I Citation2016). Bolbitis multipinna is endemic to Yunnan, China, and was first described by Wang et al. (Citation2008). It presents significant morphological variability, with one to more reticulate veins appearing on simple pinnate leaves (Zhang et al. Citation2013). Despite recent study of Bolbitis, there is little information on the chloroplast genomes (Chao et al. Citation2019). An in-depth analysis of chloroplast genomics of this genus will contribute to the development of a phylogenetic framework for morphological and taxonomical studies of the species.
The fresh leaves of B. multipinna were collected in Xishuangbanna County, Yunnan, China (101°32′28.32″E, 21°87′36.54″N, altitude: 750 m) (), and the specimen (no. WFG5594-13) has been deposited at the South China Botanical Garden Herbarium (http://herbarium.scbg.cas.cn/, contact person: Feiyan Zeng, and email: [email protected]). The genomic DNA in leaf tissue samples was extracted using CTAB following the manufacturers’ protocols (Doyle Citation1987), and sequenced by Illumina Hiseq X-Ten platform (Illumina Inc., San Diego, CA). As a result, the sequences were assembled using the GetOrganelle 1.7.5 toolkit, which depends on SPAdes 3.11 and bowtie2 (Bankevich et al. Citation2012; Langmead and Salzberg Citation2012; Jin et al. Citation2020). Annotation of complete chloroplast genome was done by using the online tool CpGAVAS2 (Shi et al. Citation2019) and submitted to GenBank under accession number OM891302.
Figure 1. The species reference image for B. multipinna. (A) Plant shape of B. multipinna. (B) Morphological characteristics of leaves of B. multipinna (lobes varied in length and irregular, reticulate veins conspicuous). The species photo was taken by the author in Mengla County, Xishuangbanna, Yunnan, China, November 2019, without any copyright issues.

The raw reads of B. multipinna are approximately 6 G, and there are 152,437 bp in the B. multipinna chloroplast genome, including a large single-copy (LSC) region of 82,618 bp, a small single-copy (SSC) region of 21,403 bp, and a pair of inverted repeat regions of 24,208 bp (IRa and IRb). In total, 132 genes are encoded by the B. multipinna chloroplast genome, including 89 protein-coding genes, 35 tRNA genes, and eight rRNA genes (). In the LSC and SSC regions as well as the IRs, the GC content was 41.3%, 38.8%, and 45.3%, respectively.
In order to infer the phylogenetic position of the newly sequenced B. multipinna in Bolbitis, we constructed a phylogeny using this newly sequenced genome in conjunction with 14 published chloroplast genomes in NCBI (). The 71 common chloroplast protein-coding genes were extracted from the 15 complete genomes, which were then aligned using MAFFT v7 (Katoh and Standley Citation2013). By concatenating these alignments, phylogenetic trees were generated using maximum-likelihood (ML) and the Bayesian inference (BI) methods in IQ-TREE v1.4.2 (Nguyen et al. Citation2015) and MrBayes v3.2.1 (Ronquist et al. Citation2012). Based on the results, B. multipinna was found to be more closely related to B. heteroclita with strong support (MLBS: 100%, BIPP: 1). Bolbitis was well supported as the sister species of Elaphoglossum (MLBS: 100%, BIPP: 1), which is in agreement with previous studies (Moran Citation2010; Liu et al. Citation2016).
Author contributions
The paper was drafted by Xinxin Cheng who assembled and annotated the samples; data analysis and interpretation were performed by Liyun Nie and Yujie Liao; Faguo Wang was responsible for the experimental design and the revision of the paper; all authors are accountable for all aspects of the work.
Ethics statement
Neither the samples used in this study nor the sampling site is listed on either the IUCN Red List of Threatened Species or the List of State-protected Plant Species. Both field and laboratory studies were conducted in accordance with the guidelines formulated by the South China Botanical Garden, Chinese Academy of Sciences.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data of this study are available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under accession no. OM891302. The associated BioProject, SRA, and BioSample numbers are PRJNA894320, SRR22044602, and SAMN31452502, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chao YS, Huang YF, Dong SY, Huang YM, Liu HY. 2019. Bolbitis lianhuachihensis (Dryopteridaceae), a new species from Taiwan. PhytoKeys. 131:69–81.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DE. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):41.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Liu HM, Zhang XC, Wang MP, Shang H, Zhou SL, Yan YH, Wei XP, Xu WB, Schneider H. 2016. Phylogenetic placement of the enigmatic fern genus Trichoneuron informs on the infra-familial relationship of Dryopteridaceae. Plant Syst Evol. 302(3):319–332.
- Moran RC, Labiak PH, Sundue M. 2010. Phylogeny and character evolution of the bolbitidoid ferns (Dryopteridaceae). Int J Plant Sci. 171(5):547–559.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- PPG I. 2016. A community-derived classification for extant lycophytes and ferns. J Syst Evol. 54:563–603.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acid Res. 47(1):65–73.
- Wang FG, Imatsuki K, Xing FW. 2008. A new name of Bolbitis from China. Am Fern J. 98(2):96–97.
- Zhang LB, Wu SG, Xiang JY, Xing FW, He H, Wang FG, Lu SG, Dong SY, Barrington DS, Iwatsuki K, et al. 2013. Dryopteridaceae. In: Wu ZY, Raven PH, editors. Flora of China. Vols. 2–3. Beijing; St. Louis: Science Press and Missouri Botanical Garden Press; p. 713–720.