Abstract
In this study, we assembled and characterized the complete chloroplast (cp) genome of Angelonia angustifolia Benth., 1846, a herbaceous and perennial plant, native to Latin America. It is an ornamental and medicinal plant that showed bright prospects for application. The cp genome of A. angustifolia has a typical conserved quadripartite structure of 154,316 bp in total length. The genome includes a large single-copy (LSC) region (84,110 bp), a small single-copy (SSC) region (15,950 bp), and a pair of inverted repeat (IR) regions (27,128 bp). The cp genome contains 130 genes comprising 85 protein-coding, 37 tRNA, and 8 rRNA genes. Phylogenetic analysis indicates that A. angustifolia is closely related to Bacopa monnieri, Scoparia dulcis, and Limnophila sessiliflora in the Plantaginaceae. Taken together, the complete cp genomes of A. angustifolia provided significant insights and important information for molecular biology, evolution, and taxonomy in the genus Angelonia.
There are now more than 26 species in the genus Angelonia, many of which are utilized to study plant modeling, flower form, colors, and floral scent volatiles (Martins et al. Citation2014; Katsu et al. Citation2017; Ghissing and Mitra Citation2021). One of these species, Angelonia angustifolia Benth. (first mentioned in 1846, https://florida.plantatlas.usf.edu/) is a herbaceous, perennial plant native to Latin America (Deyrup et al. Citation2014). This species is resistant to heat temperature but is cold-sensitive (Seaton et al. Citation2014). This species is well-known for its high ornamental and medicinal value (Deyrup et al. Citation2014). Angelonia angustifolia is commonly used as potted plant for landscaping (Blanchard and Runkle Citation2011; Deyrup et al. Citation2014). It contains numerous medicinal active ingredients such as anti-inflammatory metabolites (Deyrup et al. Citation2014; Katsu et al. Citation2017). However, the systematic position of the genus Angelonia was uncertain (APG Citation2003, Citation2009). Over the past several decades, the genus Angelonia has been placed in the Scrophulariaceae family and Plantaginaceae family (Vogel and Machado Citation1991; Martins et al. Citation2014). The chloroplast (cp) genome has been used to investigate the developmental and phylogenetic information of plants because of its maternal inheritance and conserved structure (Wang et al. Citation2018). In this present study, the complete cp genome of A. angustifolia was assembled and analyzed to better understand the phylogenetic position of A. angustifolia.
Angelonia angustifolia leaf specimens were sampled from Xinyang, Henan Province, China (the experimental base of Xinyang Agriculture and Forestry University: 114°12′ E, 32°16′ N, altitude: 102 m). Afterwards, these specimens (Bio-sample accession: SAMN25210007) were stored at −80°C of the Horticultural Plant Biotechnology Laboratory. A specimen was deposited at the Herbarium of the Horticultural Plant Biotechnology Laboratory, Xinyang Agriculture and Forestry University (contact Jianhua Yue, [email protected]) under the voucher code XAA2201102. Genomic DNA was extracted by the CTAB method (Odahara et al. Citation2009). After the DNA extraction from leaf tissues, the DNA sample was sent to Shanghai Origingene Biotechnology Co., Ltd. (Shanghai, China) to construct a DNA library. Then, the DNA library was sequenced by using the Illumina NovaSeq 6000 sequencing platform (Illumina, San Diego, CA, USA). Approximately, 7.8 GB of raw data was generated with 150 bp paired-end read lengths. Raw reads in fastq format were filtered using Trimmomatic v0.39 with default parameters retrieving the clean data (Bolger et al. Citation2014). The data were de novo assembled using NOVOPlasty based on clean reads (Dierckxsens et al. Citation2016). Then, to check the accuracy of assembly results, the slimmed assembly graph and selected target assembly graph were visualized by Bandage (Wick et al. Citation2015). The cp genome annotation was performed by Geneious v 11.1.5 (Biomatters, Auckland, New Zealand) (Kearse et al. Citation2012).
The complete cp genome of A. angustifolia has a typical conserved quadripartite structure of 154,316 bp in length with an overall GC content of 37.66%, containing four distinct regions: an large single-copy (LSC) region (84,110 bp), an small single-copy (SSC) region (15,950 bp), and a pair of inverted repeat (IR) regions (27,128 bp). The complete cp genome consists of 130 genes, comprising 85 protein coding, 37 tRNA, and 8 rRNA genes. The average coverage of the cp genome reached ×27,297 sequencing depth.
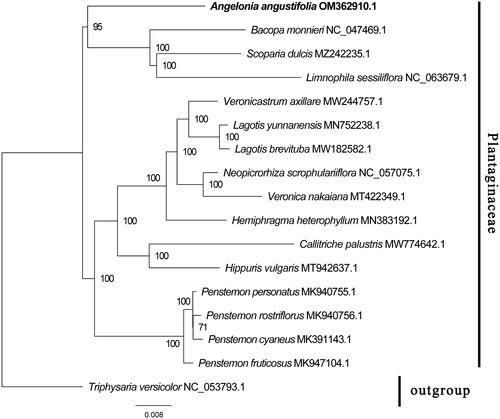
To explore the phylogenetic relationship of A. angustifolia with its closely related species, a phylogenetic tree was constructed based on the cp genome of 16 species which were downloaded from NCBI GenBank. The sequences were aligned by MAFFT v7.307 using regular settings (Katoh and Standley Citation2013), and MEGA X was used to construct the phylogenetic tree (Kumar et al. Citation2018). The robustness of the topology was calculated using the maximum-likelihood method. The program operating parameters were set as follows: a Tamura-Nei nucleotide substitution model with 1000 bootstrap replicates, accompanied by Gamma distributed with Invariant site (G + I) rates, and partial deletion of gaps/missing data according to Nguyen et al. (Citation2015). Phylogenetic analysis results strongly supported that A. angustifolia was fully resolved in a clade with Bacopa monnieri, Scoparia dulcis, and Limnophila sessiliflora in the Plantaginaceae family (). The analysis of the cp genome of A. angustifolia may provide a theoretical basis for genetic breeding, as well as determining phylogenetic relationships of A. angustifolia with related species.
Ethical approval
Plant material collection complied with the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. The permissions of plant material collection were not required as this is a popular cultivated species and is abundantly available worldwide.
Author contributions
JY and SG conceptualized and designed the research. SG provided the material for sequencing. YD collected sample. JY analyzed data and wrote the manuscript. YT validated data and modified the phylogenetic tree. YT and SG revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
The authors wish to thank the anonymous reviewers who provided constructive comments and critical insight on this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/. The complete cp genome has been deposited in GenBank under the accession no. OM362910. The associated Bio-project, SRA, and Bio-sample numbers are PRJNA799816, SRX13877377, and SAMN25210007, respectively.
Additional information
Funding
References
- APG. 2003. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 141(4):399–436.
- APG. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161(2):105–121.
- Blanchard MG, Runkle ES. 2011. Quantifying the thermal flowering rates of eighteen species of annual bedding plants. Sci Hortic. 128(1):30–37.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Deyrup ST, Asghar KB, Chacko A, Hebert JM, Samson E, Talone CJ. 2014. Chemical investigation of the medicinal and ornamental plant Angelonia angustifolia Benth. reveals therapeutic quantities of Lupeol. Fitoterapia. 98:174–178.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Ghissing U, Mitra A. 2021. Biology of floral scent volatiles in ornamental plants. In: Datta SK, Gupta YC, editors. Floriculture and ornamental plants. Handbooks of crop diversity: conservation and use of plant genetic resources. Singapore: Springer.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Katsu K, Suzuki R, Tsuchiya W, Inagaki N, Yamazaki T, Hisano T, Yasui Y, Komori T, Koshio M, Kubota S, et al. 2017. A new buckwheat dihydroflavonol 4-reductase (DFR), with a unique substrate binding structure, has altered substrate specificity. BMC Plant Biol. 17(1):239.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Martins AC, Scherz MD, Renner SS. 2014. Several origins of floral oil in the Angelonieae, a southern hemisphere disjunct clade of Plantaginaceae. Am J Bot. 101(12):2113–2120.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Odahara M, Kuroiwa H, Kuroiwa T, Sekine Y. 2009. Suppression of repeat-mediated gross mitochondrial genome rearrangements by RecA in the moss Physcomitrella patens. Plant Cell. 21(4):1182–1194.
- Seaton K, Bettin A, Grüneberg H. 2014. New ornamental plants for horticulture. In: Dixon G, Aldous D, editors. Horticulture: plants for people and places. Vol. 1. Dordrecht: Springer.
- Vogel S, Machado IC. 1991. Pollination of four sympatric species of Angelonia (Scrophulariaceae) by oil-collecting bees in NE Brazil. Plant Syst Evol. 178(3–4):153–178.
- Wang J, Li C, Yan C, Zhao X, Shan S. 2018. A comparative analysis of the complete chloroplast genome sequences of four peanut botanical varieties. PEERJ. 6:e5349.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.