Abstract
We describe the first time sequencing and assembly of the complete mitochondrial genome of Macromia manchurica Asahina, 1964 (Odonata; Macromiidae; Macromia). The mitochondrial genome of M. manchurica was found to be 15,560 bp. It contains thirteen protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and AT-rich region. The overall base composition of A. japonicus is A-38.6%, C-17.0%, G-12.5%, and T-31.9%. A phylogenetic analysis of 14 species within the order Odonata and order Ephemeroptera suggested that Macromia amphigena is most closely related to M. manchurica.
Introduction
Macromiidae belongs to the Odonata and consists of 125 species (Dijkstra et al. Citation2013; Greiner et al., Citation2019). Macromia manchurica (Odonata, Macromiidae) had been classified as a genus of Macromia (Lieftinck Citation1955; ). This species is widely distributed in Korea, Japan, China and Russia. In the genus macromia distributed in Korea, it is known as Macromia amphigena, M. manchurica, Macromia daimoji. Currently, only the mitochondrial complete genome sequence of M. amphigena and M. daimoji have been reported (Kim et al. Citation2018; An et al. Citation2022). Also we report the complete mt genome of M. manchurica to provide genetic information that can be used for various studies in the future.
Figure 1. Macromia manchurica Asahina, 1964 (Odonata; Macromiidae; Macromia) larva image. Unlike M. amphigena larva, there are no setae on the legs.

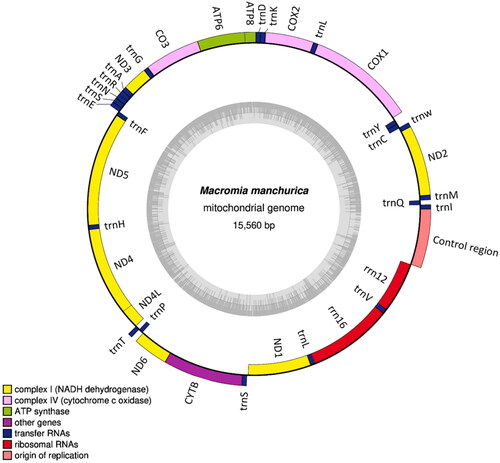
Figure 2. Mitochondrial genome map of the Macromia manchurica. A circular mitochondrial genome map was drawn using OGDRAW program (Greiner et al. Citation2019).
Materials and methods
Macromia manchurica specimen (voucher number: SJAEMM001; HG Lee, [email protected]) used in mitochondrial genome analysis was collected from Yeongokcheon, Yeongok-myeon, Gangneung-si, Gangwon-do (37°50′47.81″N, 128°48′26.01″E). The collected specimen were stored in the Department of Biological Science of Sangji University in Korea. Ethics approval for this study was obtained from the institutional Ethics Committee of Sangji University. We did not collect material from any privately owned or protected area that required permission. Total genomic DNA was extracted from the specimen using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). Genome sequencing was performed on MiSeq (Illumina Inc., San Diego, CA) platform. The treatments of raw data were performed by using Geneious prime 2022.2.1 (Biomatters Ltd, Auckland, New Zealand). To reveal the phylogenetic position of M. manchurica, 13 species downloaded and their GenBank accession numbers are provided in . We also compared each gene to the previously published mitochondrial genome of M. amphigena (MZ504971), which was suggested to be the most closely related species (An et al. Citation2022). Isonychia ignota and Ephemera orientalis were used as the outgroups. The sequences were aligned using MAFFT (Katoh et al. Citation2002) and the maximum likelihood (ML) and neighbor joining (NJ) trees were made by MEGA X (Kumar et al. Citation2018).
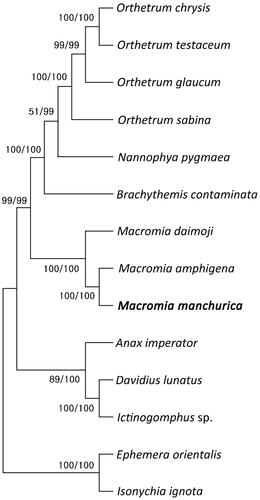
Figure 3. Phylogenetic tree of 12 order Odonata including M. manchurica and two outgroup species in order Ephemeroptera. Reconstruction of maximum likelihood (ML) and neighbor joining (NJ) trees was based on 15 genes (13 PCGs, two rRNA). Numbers at the branches represent the bootstrap support values for ML (left) and NJ (right), respectively. Branching patterns and branch lengths follow the results of ML analysis. The following sequences were used: O. Chrysis KU361233 (Yong et al. Citation2016), O. testaceum KU361235 (Yong et al. Citation2016), O. glaucum KU361232 (Yong et al. Citation2016), O. sabina KU361234 (Yong et al. Citation2016), N. pygmaea KY402222 (Jeong et al. Citation2017), B. contaminata KM658172 (Yu et al. Citation2016), M. daimoji NC_041425 (Kim et al. Citation2018), M. amphigena MZ504971 (An et al. Citation2022), A. imperator KX161841 (Herzog et al. Citation2016), D. lunatus EU591677 (Lee et al. Citation2009), Ictinogomphus sp. KM244673 (Tang et al. Citation2014), E. orientalis EU591678 (Lee et al. Citation2009), I. ignota HM143892 (unpublished).
Results
The assembled mitogenome of M. manchurica (GenBank accession No: MZ504972) showed a length of 15,560 bp. Also, the total GC content is 16.8% and the total nucleotide composition is A − 38.6%, C − 17.0%, G − 12.5%, and T − 31.9%. The mitogenome consists of thirteen protein-coding genes (PCGs), 22 tRNA genes, and two ribosomal RNA (rRNA) genes (). The genome structure, gene order, and total gene number of M. manchurica mt genome is identically same to M. amphigena (MZ504971), which is closely related genus. The two independent phylogenetic trees yielded the same topology. Macromia manchurica formed a monophyly with M. amphigena with a high support value (BS = 100) ().
Conclusion
The mitochondrial genome of M. manchurica was 15,560 bp and contained thirteen protein-coding genes, 22 transfer RNAs, two ribosomal RNAs and AT-rich region. A phylogenetic analysis of fourteen complete mitochondrial genomes of the registered Order Odonata and Order Ephemeroptera suggested that M. amphigena was most closely related to M. manchurica. Also, among the genus macromia inhabiting Korea, the M. manchurica was more closely related to the M. amphigena than the M. daimoji. We expect that the present results will facilitate further investigations into the phylogenetic relationship of the genus Macromia.
Author contributions
Kyeong-Sik Cheon was involved in the conception and design; Chae-Hui An was involved in the analysis and interpretation of the data; Chae-Hui An and Ji-Eun Jang was involved in the DNA extraction experiment; Chae-Hui An and Hwang-Goo Lee drafted the paper; all authors revised it critically for intellectual content and gave the final approval of the version to be published; and that all authors agree to be accountable for all aspects of the work.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. under the accession no. MZ504972. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA744014, SRR15092394 and SAMN20065584, respectively.
Additional information
Funding
References
- An CH, Cheon KS, Jang JE, Choi JK, Lee HG. 2022. Complete mitochondrial genome of large dragonfly (Macromia amphigena). Mitochondrial DNA B Resour. 7(2):377–378.
- Dijkstra KDB, Bechly G, Bybee SM, Dow RA, Dumont HJ, Fleck G, Garrison RW, Hämäläinen M, Kalkman VJ, Karube H, et al. 2013. The classification and diversity of dragonflies and damselflies (Odonata). Zootaxa. 3703(1):36–45.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- Herzog R, Osigus HJ, Feindt W, Schierwater B, Hadrys H. 2016. The complete mitochondrial genome of the emperor dragonfly Anax imperator LEACH, 1815 (Odonata: aeshnidae) via NGS sequencing. Mitochondrial DNA B Resour. 1(1):783–786.
- Jeong SY, Kim MJ, Wang AR, Kim SS, An J, Kim I. 2017. Complete mitochondrial genome sequence of the tiny dragonfly, Nannophya pygmaea (Odonata: Libellulidae). Conserv Genet Reour. 10:355–358.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kim MJ, Jeong SY, Wang AR, An JH, Kim IS. 2018. Complete mitochondrial genome sequence of Macromia daimoji Okumura, 1949 (Odonata: macromiidae). Mitochondrial DNA B Resour. 3(1):365–367.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee EM, Hong MY, Kim MI, Kim MJ, Park HC, Kim KY, Lee IH, Bae CH, Jin BR, Kim I. 2009. The complete mitogenome sequences of the palaeopteran insects Ephemera orientalis (Ephemeroptera: ephemeridae) and Davidius lunatus (Odonata: gomphidae). Genome. 52(9):810–817.
- Lieftinck MA. 1955. Further inquiries into the Old World species of Macromia rambur (Odonata). Zoologische Mededelingen. 33(25):251–277.
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes - a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42(22):e166.
- Yong HS, Song SL, Suana IW, Eamsobhana P, Lim PE. 2016. Complete mitochondrial genome of Orthetrum dragonflies and molecular phylogeny of Odonata. Biochem Syst Ecol. 69:124–131.
- Yu P, Cheng X, Ma Y, Yu D, Zhang J. 2016. The complete mitochondrial genome of Brachythemis contaminate (Odonata: libellulidae). Mitochondrial DNA Part A. 27:2272–2273.
