Abstract
The catfish, Pangasius nasutus and P. conchophilus, are often misidentified between each other due to their similar morphology. Thus, the current study was conducted to differentiate them based on a molecular approach. The complete mitochondrial genomes of P. nasutus and P. conchophilus obtained from the Pahang River (Peninsular Malaysia) were sequenced, assembled, and annotated using next-generation sequencing (NGS). A 16,465 bp and 16,470 bp length mitogenome sequence of P. nasutus and P. conchophilus, respectively, was generated, each containing 13 protein genes, 22 tRNAs, and two rRNAs, typical of most vertebrates. This is the first report of the complete mitochondrial genome sequences of P. nasutus and P. conchophilus. These data are a valuable genetic resource for future studies of these two commercially important species.
Introduction
Pangasius nasutus (Bleeker Citation1863) is a commercially important species of catfish in the genus Pangasius. In Malaysia, it is locally referred to as 'Patin buah’. It has a disjunctive distribution occurring in Pahang River and its tributaries (Peninsular Malaysia), Batang Rajang (Malaysian Borneo), Batang Hari, Indragiri, Musi (Sumatra) and Kalimantan, Barito, Kahayan, Kapuas (Indonesian Borneo) (Roberts and Vidthayanon Citation1991; Parenti and Lim Citation2005; Kottelat Citation2013; Gustiano Citation2016). It is a native species in the Pahang River, the largest river in Malaysia. Pangasius nasutus is differentiated from all other species in the genus Pangasius by having an inferior mouth type, an entirely exposed tooth band of the upper jaw when the jaws are closed, and a strongly projected snout (Roberts and Vidthayanon Citation1991). Pangasius conchophilus Roberts and Vidthayanon Citation1991, is locally referred to as 'Patin buah kemboja’, and is also considered a commercially important species of catfish in the genus Pangasius and constantly misidentified with P. nasutus. Unlike the native P. nasutus, P. conchophilus is an introduced species into Malaysia. Its native origin is from the Mekong, Bangpakong, and Chao Phraya basins (Roberts and Vidthayanon Citation1991; Kottelat Citation2013). During the 1990s, P. conchophilus fish fries were brought in from Cambodia through Thailand and farmed along the Pahang River by Cambodian immigrants (Baharuddin Citation2016). Pangasius conchophilus is differentiated from all other species in the genus Pangasius by having a subterminal mouth, less strongly projecting snout, and large eye diameter (Roberts and Vidthayanon Citation1991; Baharuddin Citation2016). Several authors have recognized a very close relationship between P. nasutus and P. conchophilus and they often misidentified as each other (Roberts and Vidthayanon Citation1991; Pouyaud et al. Citation1998; Pouyaud et al. Citation2000; Baharuddin Citation2016; Pouyaud et al. Citation2016; Gustiano et al. Citation2018). To our knowledge, there is no previous study on the mitogenomes of these two species, which raises questions about their evolutionary relationships. To better understand the taxonomic relationships between the two species, we sequenced, assembled, and annotated the whole mitochondrial genome of P. nasutus and P. conchophilus, which would serve as a significant genomic resource for future research.
Materials
A single adult specimen of P. nasutus (specimen ID: TM15) () was collected from Kuala Krau, Temerloh River (3°42′25.9″N 102°22′25.3″E), while the P. conchophilus individual (specimen ID: JR15) () was sampled from Kuala Tembeling, Jerantut River (4°04′13.4″N 102°18′57.1″E). Both Temerloh and Jerantut rivers were part of the Pahang River tributaries. The fish specimens were morphologically identified based on Roberts and Vidthayanon (Citation1991), Kottelat (Citation2013), and Baharuddin (2016). The fish muscle from the caudal peduncle and pectoral fins were excised, preserved in 99% ethanol, and stored at −80 °C until further use for DNA extraction. All specimens are deposited in the Depository Museum, Department of Aquaculture, Faculty of Agriculture, Universiti Putra Malaysia (Dr Zafri Hassan, +60397694932, [email protected]) under the voucher number JAQ/PN/00001 and JAQ/PC/00001.
Methods
Approximately, 20–50 mg of tissue sample was used for DNA extraction using WizPrep gDNA Mini Kit (WizBio, Seongnam, South Korea) based on the manufacturer’s instructions. DNA samples were treated with 1 μL of RNAse (10 mg/mL) for 30 min at room temperature, followed by purification using a 1× volume of SPRI bead (Oberacker et al. Citation2019). Then, 2 μL of the purified DNA was measured using Denovix high sensitivity kit (Denovix, Wilmington, DE). Approximately, 100 ng of DNA was fragmented to 350 bp using a Bioruptor followed by NEB Ultra II library preparation according to the manufacturer’s instructions (NEB, Ipswich, MA). Sequencing was performed on a NovaSEQ6000 (Illumina, San Diego, CA) using a run configuration of 2 × 150 bp to generate approximately 1 Gb of data for each sample. Raw reads were trimmed with fastp v0.21 (https://github.com/OpenGene/fastp.git) (Chen et al. Citation2018) to remove low-quality bases and Illumina adapter sequences. The trimmed reads were subsequently used for de novo assembly in MegaHIT (default setting) (https://github.com/voutcn/megahit.git) (Li et al. Citation2015). The mitochondrial-derived contigs were identified, circularized, and annotated using MitoZ (https://github.com/linzhi2013/MitoZ.git) (Meng et al. Citation2019).
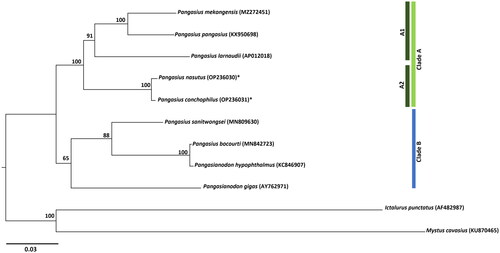
To investigate the evolutionary relationships among the Pangasius species, the mitogenomes of P. nasutus and P. conchophilus were aligned with the seven available Pangasiid species mitogenome sequences from GenBank using MUSCLE as implemented in MEGA X (Kumar et al. Citation2018). The maximum-likelihood (ML) method was employed for phylogenetic reconstruction of the Pangasiid species to determine their relationships. Ictalurus punctatus (family Ictaluridae) and Mystus cavasius (family Bagridae) were selected as the outgroups. Considering all positions, the ML phylogenetic tree was calculated under the general time reversible (GTR) model using the software RAxML-NG (Kozlov et al. Citation2019) executed in the graphical interface raxmlGUI 2.0 (Edler et al. Citation2021). To evaluate the robustness of each node, the bootstrap proportions were computed (500 replicates). The generated tree is displayed in FigTree v1.4.4 (Rambaut Citation2018).
Results
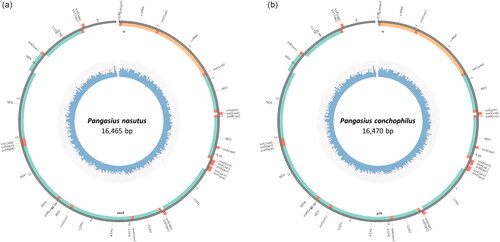
Both complete mitogenome sequences have been deposited at the National Center for Biotechnology Information GenBank (NCBI) database under the accession numbers OP236030 (Pangasius nasutus) () and OP236031 (P. conchophilus) (). Each mitogenome consists of 37 genes; 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes (duplication of two tRNAs: tRNALeu and tRNASer) on both strands, and two ribosomal RNA (rRNA) genes. The complete mitogenome of P. nasutus is a circular molecule with a length of 16,465 bp, and a nucleotide composition of A: 31.3%, T: 26.1%, G: 15.1%, and C: 27.5%. Of the 13 PCGs, only the COX1 gene starts with GTG, while the other 12 genes originate from ATG, in parallel with the other Pangasiid fishes (Jondeung et al. Citation2007; Wei et al. Citation2020; Ni et al. Citation2021). Only Cytb contains an incomplete stop codon (T), while the other genes end with a complete stop codon (TAA or TAG). The complete mitogenome of P. conchophilus is 16,470 bp in length, with a nucleotide composition of A: 31.2%, T: 26.1%, G: 15.1%, and C: 27.6%. Similar to P. nasutus, of the 13 PCGs, only the CO1 gene starts with GTG, while the remaining 12 genes start with ATG. Furthermore, all PCGs genes contain a complete stop codon (TAA or TAG) except Cytb which contains an incomplete stop codon (T). Based on whole mitogenome alignment, the pair-wise nucleotide similarity between P. nasutus and P. conchophilus is 0.6%.
The phylogenetic study revealed two main clusters (A and B) (). The first cluster bifurcated into two minor clusters (A1 and A2). Pangasius larnaudii is closely related to P. mekongensis and P. pangasius, which is supported by previous research (Ni et al. Citation2021). In the second minor cluster (A2) Pangasius nasutus and P. conchophilus form another closely related species and sister to cluster A. The second main cluster (B) shows the close relationship of P. bocourti and Pangasianodon hypophthalmus forming a sister cluster to P. sanitwongsei. Pangasianodon gigas is sister to this cluster in main cluster B, consistent with prior findings (Kim et al. Citation2018; Chen et al. Citation2020). Most major and minor clusters are well supported (BP: 100%).
Discussion and conclusions
We present here the first report of the complete mitochondrial genomes of Pangasius nasutus and P. conchophilus, having successfully sequenced, assembled, and annotated. The complete structure of the mitogenome published here could be used as a foundation for further research into the population genetics, evolutionary biology, phylogenetic analysis, aquaculture, genomics as well as species identification of Pangasiid catfish and related species.
Author contributions
Siti Amalia Aisyah Abdul Halim: conceptualization, methodology, formal analysis, investigation, writing – original draft, review and editing. Yuzine Esa: conceptualization, methodology, investigation, writing –review and editing, funding acquisition. Han Ming Gan: formal analysis, investigation, writing – review and editing. Amir Asyraf Zainudin: resources, investigation, writing – review and editing. Siti Azizah Mohd Nor: resources, investigation, writing – review and editing. All authors agreed to be accountable for all aspects of the work and approved the final draft to be published.
Ethics statement
The study protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the Office of the Deputy Vice Chancellor (Research & Innovation), Universiti Putra Malaysia, Selangor, Malaysia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete mitogenome sequences of Pangasius nasutus and P. conchophilus have been deposited in the NCBI GenBank database at https://www.ncbi.nlm.nih.gov/ under the accession numbers OP236030 and OP236031, respectively. The associated BioProject, SRA, and Bio-Sample numbers are Pangasius nasutus, PRJNA891653, SRR21979525, and SAMN31373448 and for P. conchophilus, PRJNA891658, SRR21979584, and SAMN31373450, respectively.
Additional information
Funding
References
- Baharuddin H. 2016. Morphometric, phylogenetic analyses and DNA barcoding of Pangasiid catfishes (Teleostei: Pangasiidae) in Peninsular Malaysia [PhD dissertation]. Kuala Lumpur: Universiti Malaya.
- Bleeker P. 1863. Atlas ichthyologique des Indes Orientales Néêrlandaises. Tome II. Siluroïdes, chacoïdes et hétérobranchoïdes. Amsterdam: Müller.
- Chen J, Gao T, Chen M, Ou Q. 2020. Next-generation sequencing of the mitochondrial genome of Pangasius bocourti (Siluroidei: Pangasiidae). Mitochondrial DNA B Resour. 5(2):1779–1780.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Edler D, Klein J, Antonelli A, Silvestro D. 2021. raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol Evol. 12(2):373–377.
- Gustiano R. 2016. Pangasiid catfishes of Indonesia. BPN. 15(2):91–100.
- Gustiano R, Prakoso VA, Ath-Thar MHF. 2018. Asian catfish genus Pangasius: diagnosis and distribution. Indones Fish Res J. 24(2):99–115.
- Jondeung A, Sangthong P, Zardoya R. 2007. The complete mitochondrial DNA sequence of the Mekong giant catfish (Pangasianodon gigas), and the phylogenetic relationships among Siluriformes. Gene. 387(1–2):49–57.
- Kim OTP, Nguyen PT, Shoguchi E, Hisata K, Vo TTB, Inoue J, Shinzato C, Le BTN, Nishitsuji K, Kanda M, et al. 2018. A draft genome of the striped catfish, Pangasianodon hypophthalmus, for comparative analysis of genes relevant to development and a resource for aquaculture improvement. BMC Genomics. 19(1):733.
- Kottelat M. 2013. The fishes of the inland waters of Southeast Asia: a catalogue and core bibliography of the fishes known to occur in freshwaters, mangroves and estuaries. Raffles Bull Zool Suppl. 27:1–663.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 31(10):1674–1676.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):1–7.
- Ni W, Liu Y, Chen H, Yu L, Li W, Wang Y, Hong X, Chen C, Yuan J, Liu F, et al. 2021. The complete mitochondrial genome of Pangasianodon hypophthalmus (Sauvage 1878) (Siluriformes, Pangasiidae). Mitochondrial DNA B Resour. 6(12):3391–3392.
- Oberacker P, Stepper P, Bond DM, Höhn S, Focken J, Meyer V, Schelle L, Sugrue VJ, Jeunen G-J, Moser T, et al. 2019. Bio-On-Magnetic-Beads (BOMB): open platform for high-throughput nucleic acid extraction and manipulation. PLoS Biol. 17(1):e3000107.
- Parenti LR, Lim KKP. 2005. Fishes of the Rajang Basin, Sarawak, Malaysia. Raffles Bull Zool. 13:175–208.
- Pouyaud L, Teugels GG, Gustiano R, Legendre M. 2000. Contribution to the phylogeny of Pangasiid catfishes based on allozymes and mitochondrial DNA. J Fish Biol. 56(6):1509–1538.
- Pouyaud L, Gustiano R, Legendre M. 1998. Phylogenetic relationships among Pangasiid catfish species (Siluriformes, Pangasiidae) and new insights on their zoogeography. In: Legendre M, Pariselle A, editors. The biological diversity and aquaculture of clariid and pangasiid catfishes in South-East Asia: proceedings of the mid-term workshop of the "Catfish Asia project". Vietnam: Cantho; p. 49–56.
- Pouyaud L, Gustiano R, Teugels GG. 2016. Contribution to the phylogeny of the Pangasiidae based on mitochondrial 12S RDNA. Indones J Agric Sci. 5(2):4562–4562.
- Rambaut A. 2018. FigTree: tree figure drawing tool v1.4.4. https://github.com/rambaut/figtree/releases.
- Roberts TR, Vidthayanon C. 1991. Systematic revision of the Asian catfish family Pangasiidae, with biological observations and descriptions of 3 new species. Proc Acad Nat Sci Philadelphia. 143:97–144.
- Wei L, Ye X, Lv Y, Teng Z, Gan B, Zou H, Mo F, Zhang S. 2020. Complete mitochondrial genome and phylogenetic position of Pangasius sanitwongsei (Siluriformes: Pangasiidae). Mitochondrial DNA B Resour. 5(1):945–946.