Abstract
Grateloupia elliptica (Holmes, 1896) is a red alga belonging to the order Halymeniales and phylum Rhodophyta. In this study, the complete mitochondrial DNA (mtDNA) of G. elliptica has been described. The complete circular mtDNA of G. elliptica was 28,503 bp in length, with an A + T content of 68.78%; it encoded a total of 49 genes, including 20 tRNA, three rRNA, and 26 protein-coding (CDS) genes. Phylogenetic analysis based on complete mitochondrial genomes revealed that G. elliptica was most closely related to G. angusta. The complete mitochondrial sequence of G. elliptica will enrich the mitochondrial genome database and provide useful resources for population genetics and evolution analyses.
Grateloupia elliptica is a red marine macroalga belonging to the genus Grateloupia (Halymeniaceae; Halymeniales; Florideophyceae; Rhodophyta). G. elliptica is commonly referred to as an intertidal alga and is found across Korea and Japan (Yang et al. Citation2013). Recent research has suggested that bioactive substances found in G. elliptica may have medicinal benefits (Lee et al. Citation2021). Grateloupia is the largest genus in the Halymeniaceae family, with several species that are difficult to distinguish (Miller et al. Citation2011). Mitochondrial genes and full mitochondrial DNA (mtDNA) are commonly used for conducting algal species identification, population genetics, and biogeographic investigations (Lee et al. Citation2009; Kim et al. Citation2012). The current work examines the complete mtDNA of G. elliptica and its evolutionary relationship within the genus.
The G. elliptica specimen used in this study was collected from the East Sea, South Korea (37°06′41.5′′N 129°22′45.6′′E) and deposited at the Ecological Restoration Group, Marine Eco-Technology Institute, Busan, South Korea (Young-Ryun Kim, [email protected]) under the voucher number PU-T01-S-MA-02. Total DNA was extracted using DNeasy Blood and tissue kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. A DNA library was prepared using TruSeq Nano DNA Kit and sequenced on an Illumina platform (HiSeq2500, San Diego, CA); subsequently, 150 bp paired-end reads were generated. The obtained raw data were preprocessed for quality filtering and trimming of low-quality bases and adapters by FastQC. A total of 1,665,114,426 read bases of filtered data were analyzed to generate 11,126,568 reads of sequence; then De novo assembly was performed with the Platanus-allee version 2.2.2 (Kajitani et al. Citation2019) and complete mtDNA annotation was performed using the MFannot tool (https://megasun.bch.umontreal.ca/cgi-bin/mfannot/). A phylogenetic tree was constructed using the whole mitochondrial genome sequences of eight different species. Prior to phylogenetic analysis, the mitochondrial genome was aligned using ClustalW. The evolutionary position of G. elliptica was investigated by constructing a phylogenetic tree based on the complete mitochondrial genome using the maximum-likelihood (ML) method and conventional bootstraps analysis (1000 replications) in MEGA11 version 11.0.8 (Tamura et al. Citation2021).
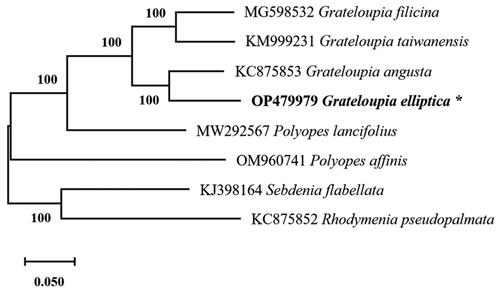
The complete circular mtDNA (GenBank accession No.: OP479979) of G. elliptica was 28,503 bp in length with 68.78% A + T content. The overall base composition was 36.20% for A (10,318 bp), 32.58% for T (9287 bp), 15.95% for G (4546 bp), and 15.27% for C (4352 bp). The mtDNA contained 49 genes, including 20 tRNA genes, three rRNA genes, and 26 protein-coding (CDS) genes. The CDS genes consisted of four ATPase subunits, six ribosomal proteins, seven NADH dehydrogenase subunits, three sdh subunits, three cox subunits, one of each cob and tatC subunits, and one intronic reading frame (ORF634). In the mtDNA of G. elliptica (OP479979), a group II intron is located within the cox1 gene, which is similar to G. angusta (Kim et al. Citation2014), G. taiwanensis (DePriest et al. Citation2014), and G. filicina (Li et al. Citation2018). The three rRNA genes have lengths of 108 bp (rrn5 rRNA), 1365 bp (rns rRNA), and 2624 bp (rnl rRNA). The phylogeny recovered a close relationship between G. elliptica with G. angusta with maximum bootstrap support (). The complete mtDNA sequence of G. elliptica may help advance our understanding of the Grateloupia evolution.
Figure 1. The phylogenetic tree (maximum likelihood) of the all publicly available Halymeniales mitogenome was included and mitogenomes from Rhodymenia pseudopalmata (Rhodymeniales) and Sebdenia flabellata (Sebdeniales) were included as outgroups. The asterisks beside Grateloupia elliptica denote the newly discovered mitochondrial genome.
Ethical approval
The marine samples collected and used for this study do not involve any marine organisms under protection determined by the Ordinance of the Ministry of Oceans and Fisheries in the Republic of Korea. Therefore, our study was exempted from ethical approval and did not need any permission to carry it out.
Authors’ contribution
M.P.P. performed the experiments, analyzed the data, was involved in certain tools for analysis, and drafting of the article, and approved the final draft. J-O.K. involved in the conception and design of the work, certain tools for analysis and sorting out of the results. Y-R.K. involved in specimen sample collection, species identification, and prepared figure. K.K. was involved in the conception and design of the work, certain tools for analysis, funding acquisition, revising it critically for intellectual content, and the final approval of the version to be published.
Disclosure statement
The authors report no conflict of interest. The authors are responsible for the content and writing of the article.
Data availability statement
The genome sequence data supporting this study’s findings are available in the GenBank database (https://www.ncbi.nlm.nih.gov/) under the accession number OP479979. The associated BioProject, BioSample, and SRA numbers are PRJNA825655, SAMN27532332, and SRR18728769, respectively.
Additional information
Funding
References
- DePriest MS, Bhattacharya D, Lopez-Bautista JM. 2014. The mitochondrial genome of Grateloupia taiwanensis (Halymeniaceae, Rhodophyta) and comparative mitochondrial genomics of red algae. Biol Bull. 227(2):191–200.
- Kajitani R, Yoshimura D, Okuno M, Minakuchi Y, Kagoshima H, Fujiyama A, Kubokawa K, Kohara Y, Toyoda A, Itoh T. 2019. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat Commun. 10(1):1–15.
- Kim KM, Hoarau GG, Boo SM. 2012. Genetic structure and distribution of Gelidium elegans (Gelidiales, Rhodophyta) in Korea based on mitochondrial cox1 sequence data. Aquat Bot. 98(1):27–33.
- Kim SY, Yang EC, Boo SM, Yoon HS. 2014. Complete mitochondrial genome of the marine red alga Grateloupia angusta (Halymeniales). Mitochondrial DNA. 25(4):269–270.
- Lee JI, Kim HG, Geraldino PJL, Hwang IK, Boo SM. 2009. Molecular classification of the genus Grateloupia (Halymeniaceae, Rhodophyta) in Korea. Algae. 24(4):231–238.
- Lee HG, Lu YA, Je JG, Jayawardena TU, Kang MC, Lee SH, Kim TH, Lee DS, Lee JM, Yim MJ, et al. 2021. Effects of ethanol extracts from Grateloupia elliptica, a red seaweed, and its chlorophyll derivative on 3T3-L1 adipocytes: suppression of lipid accumulation through downregulation of adipogenic protein expression. Mar Drugs. 19(2):91.
- Li Y, Meinita MDN, Liu T, Chi S, Yin H. 2018. Complete sequences of the mitochondrial DNA of the Grateloupia filicina (Rhodophyta). Mitochondrial DNA B Resour. 3(1):76–77.
- Miller KA, Aguilar-Rosas LE, Pedroche FF. 2011. A review of non-native seaweeds from California, USA and Baja California, Mexico. Hidrobiologica. 21(3):365–379.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027.
- Yang MY, Han EG, Kim MS. 2013. Molecular identification of Grateloupia elliptica and G. lanceolata (Rhodophyta) inferred from plastid rbcL and mitochondrial COI genes sequence data. Genes Genom. 35(2):239–246.
