Abstract
Artemisia borotalensis Poljakov is an endemic and endangered herb in China. In this study, we sequenced and analyzed the complete chloroplast genome of this species. Sequencing revealed the genome to be 151,179 bp in length, containing a large single copy region (82,862 bp), a small single copy region (18,377 bp), and a pair of inverted repeat regions (24,970 bp each). Our analyses demonstrated that it contained 133 genes, including 87 protein-coding genes, 37 transfer RNA genes, eight ribosomal RNA genes, and one pseudogene (ycf1). Furthermore, we found the genome to have an overall GC content of 37.4%. A phylogenetic analysis indicated that A. borotalensis and A. maritima clustered together as sister group to A. annua and A. fukudo clade.
Introduction
Artemisia borotalensis Poljakov Citation1954, an endemic plant in China, is a perennial herb from the family Asteraceae (Ling et al. Citation2011). In China, A. borotalensis is distributed over desert or semi-desert grassland in the Gobi and on gravel hillside areas in Xinjiang (). Most Artemisia species have important medicinal, ecological, and economic value (Duffy and Mutabingwa Citation2006; Vallès et al. Citation2011). Song et al. (2021) reported that the essential oils contained in A. borotalensis can effectively kill cotton aphids and psyllids. However, A. borotalensis has become an endangered species due to its relatively narrow area of distribution and the disturbance of its original habitat destroyed by human activities. Therefore, using the genome skimming approach, we constructed the complete chloroplast genome of A. borotalensis, which will contribute to the conservation of this taxon’s genetic resources and the understanding of its phylogenetic relationships.
Materials
An A. borotalensis specimen was collected from Xinjiang, China (46°7′25.6″ N, 84°31′58″ E; Altitude: 840 m). A. borotalensis is not a legally protected species in China, despite the decline in the size of its population. Collection of the sample followed the wild plant protection regulations of China. The voucher specimen was deposited in the Herbarium of the Institute of Botany, Chinese Academy of Sciences (PE, Xian-Chun Zhang, [email protected]), with collection number ZC-451.
Methods
Total genomic DNA was extracted from approximately 100 mg of silica-dried leaf material using a modified CTAB method (Doyle and Doyle Citation1987). DNA extracts were fragmented to construct a 300 bp short-insert library and sequenced with 2 × 150 bp paired-end reads on DNBSEQ™ technology platforms at Beijing Genomics Institute (Shenzhen, China). The raw reads were filtered using Trimmomatic 0.35 (Bolger et al. Citation2014) to remove adapters and low-quality bases. Then, about 2.5 Gb of clean reads were assembled using GetOrganelle v. 1.7.1 (Jin et al. Citation2020). The chloroplast genomes were annotated with GeSeq (Tillich et al. Citation2017), before being manually adjusted with Geneious v. 9.1.7 (Kearse et al. Citation2012). Finally, the annotated chloroplast genomes were submitted to GenBank using Bankit (Accession number: ON964524), and Organellar Genome Draw (OGDRAW) (Lohse et al. Citation2007) was used to illustrate the circular genome map.
To investigate the phylogenetic position of A. borotalensis, we reconstructed its phylogenetic relationships based on 16 complete chloroplast genomes from 10 Artemisia species and used the closely related species Ajania pacifica (NC_050690) as an outgroup. The 17 complete chloroplast genomes (after removing one inverted repeat region) were aligned using the default parameters in MAFFT v. 7 (Katoh and Standley Citation2013). According to the Bayesian information criterion (BIC), the most appropriate substitution models, estimated using jModelTest2 (Darriba et al. Citation2012), were TVM + I + G for the complete chloroplast genome sequences. Maximum likelihood (ML) analyses were then conducted using RAxML-HPC v. 8 (Stamatakis Citation2014) with 1000 bootstrap replicates. The evaluation of the most appropriate substitution models and the construction of the phylogenetic tree were both carried out on the CIPRES Science Gateway portal (Miller et al. Citation2010).
Results
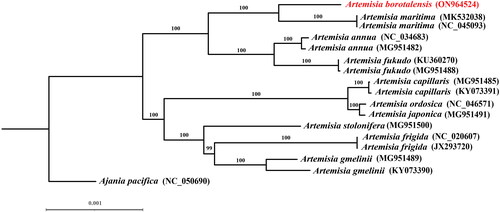
The sequenced chloroplast genome of A. borotalensis was found to possess a typical quadripartite structure, which consisted of large single copy (LSC), small single copy (SSC), and inverted repeat (IRa and IRb) regions (). The complete chloroplast genome was 151,179 bp in length, of which the LSC, SSC, and IR were 82,862 bp, 18,377 bp and 24,970 bp in length, respectively. In total, 133 genes were identified, including 87 protein-coding genes, 37 transfer RNA genes, eight ribosomal RNA genes, and one pseudogene (ycf1). The overall GC content of the chloroplast genome was shown to be 37.4%. Comparison of the A. borotalensis chloroplast genome to previously published data revealed a high level of gene synteny with one publicly available genome data sets from A. maritima (Shahzadi et al. Citation2020). Furthermore, phylogenetic analysis showed that A. borotalensis and A. maritima clustered together as sister group (with 100% bootstrap support) to A. annua and A. fukudo clade ().
Figure 2. Circular chloroplast genome map for Artemisia borotalensis Poljakov. The colored bars indicate different functional gene groups. The thick lines of the large circle indicate the extent of the inverted repeat regions (IRa and IRb) that separate the genome into small single copy (SSC) and large single copy (LSC) regions, respectively. The darker gray columns in the inner circle correspond to the guanine-cytosine (GC) content, and the light gray columns to the adenosine-thymine (AT) content.
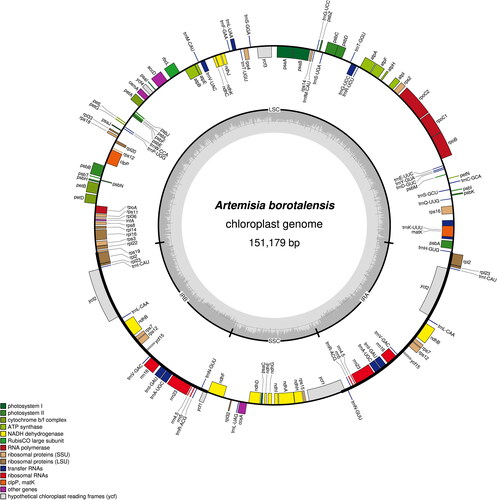
Figure 3. Phylogenetic trees inferred from maximum likelihood (ML) analyses based on 16 complete chloroplast genomes from 10 Artemisia species using Ajania pacifica as an outgroup with 1000 bootstraps replicates. The numbers above the branches indicate the bootstrap values. The species name colored in red represents our sequenced chloroplast genome (A. borotalensis). The following sequences were used: Ajania pacifica NC_050690 (Kim and Kim Citation2020), Artemisia annua MG951482 (Kim et al. Citation2020), Artemisia annua NC_034683, Artemisia capillaris KY073391 (Lee et al. Citation2022), Artemisia capillaris MG951485 (Kim et al. Citation2020), Artemisia frigida JX293720 (Liu et al. Citation2013), Artemisia frigida NC_020607, Artemisia fukudo KU360270 (Lee et al. Citation2016), Artemisia fukudo MG951488 (Kim et al. Citation2020), Artemisia gmelinii KY073390 (Lee et al. Citation2022), Artemisia gmelinii MG951489 (Kim et al. Citation2020), Artemisia japonica MG951491 (Kim et al. Citation2020), Artemisia maritima MK532038 (Shahzadi et al. Citation2020), Artemisia maritima NC_045093, Artemisia ordosica NC_046571 (Li et al. Citation2020), and Artemisia stolonifera MG951500 (Kim et al. Citation2020).
Discussion and conclusions
The chloroplast genome of A. borotalensis sequenced in this study was consistent with those of other previously published Artemisia taxa in terms of gene structure, number, and GC content (Kim et al. Citation2020), suggesting that the chloroplast genome of this taxon is relatively evolutionarily conserved. Moreover, the phylogenetic results confirmed the classification of A. borotalensis within the Artemisia subgenus Seriphidium based on its morphological traits, which has not been addressed in previous phylogenetic studies due to sampling limitations (Malik et al. Citation2017). In future, our findings will contribute to the development and conservation of the genetic resources of Artemisia and lay the foundation on which to develop a further understanding the phylogenetic relationships between members of this genus.
Author contributions
Ying Feng was mainly responsible for the study design. Guangzhao Jin conducted the analysis of data and drafted the manuscript. Zhibin Wen and Feng Song helped perform the analysis with constructive discussions and revised it critically for intellectual content. All authors approve the version to be published and agree to be accountable for all aspects of the work.
Regulation statement
All materials used in the study were collected in public areas of China and strictly follows the wild plant protection regulations of China. Voucher specimens were prepared and deposited at the Herbarium of the Institute of Botany, Chinese Academy of Sciences (PE).
Supplemental Material
Download MS Word (165.3 KB)Acknowledgments
We thank Meng Wei and Jiye Zheng of the Institute of Botany, Chinese Academy of Sciences, for their assistance in sample collection.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors have provided all the raw data, which has been all activated. The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/ under the accession number ON964524. The associated BioProject, SRA, and BioSample numbers are PRJNA857588, SRR20363929 and SAMN29630348 respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Duffy PE, Mutabingwa TK. 2006. Artemisinin combination therapies. Lancet. 367(9528):2037–2039.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z., 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):31.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim G-B, Lim CE, Kim J-S, Kim K, Lee JH, Yu H-J, Mun J-H., 2020. Comparative chloroplast genome analysis of Artemisia (Asteraceae) in East Asia: insights into evolutionary divergence and phylogenomic implications. BMC Genomics. 21(1):415.
- Kim HT, Kim JS. 2020. The complete chloroplast genome sequence of Ajania pacifica (Nakai) Bremer & Humphries. Mitochondrial DNA Part B. 5(3):2399–2400.
- Lee YS, Park JY, Kim J-K, Lee HO, Park H-S, Lee S-C, Kang JH, Lee TJ, Sung SH, Yang T-J, et al. 2016. Complete chloroplast genome sequence of Artemisia fukudo Makino (Asteraceae). Mitochondrial DNA Part B. 1(1):376–377.
- Lee YS, Woo S, Kim J-K, Park JY, Izzah NK, Park H-S, Kang JH, Lee TJ, Sung SH, Kang KB, et al. 2022. Genetic and chemical markers for authentication of three Artemisia species: A. capillaris, A. gmelinii, and A. fukudo. PLoS One. 17(3):e0264576.
- Li X, He JH, Lian YX, Li FL, Suo JH, Jin GQ. 2020. The complete chloroplast genome sequence of Artemisia ordosica. Mitochondrial DNA Part B. 5(3):2180–2181.
- Ling YR, Humphries CJ, Gilbert MG. 2011. Seriphidium (Besser ex Lessing) Fourreau. In: Wu ZY, Raven PH, editors. Flora of China, vol. 20–21. Beijing: Science Press. p. 737–747.
- Liu Y, Huo N, Dong L, Wang Y, Zhang S, Young HA, Feng X, Gu YQ., 2013. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One. 8(2):e57533.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- Malik S, Vitales D, Hayat MQ, Korobkov AA, Garnatje T, Vallès J. 2017. Phylogeny and biogeography of Artemisia subgenus Seriphidium (Asteraceae: Anthemideae). Taxon. 66(4):934–952.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE) 14 November 2010. New Orleans, LA: 1-8.
- Poljakov PP. 1954. Species novae generis Artemisia subgeneris Seriphidium (Bess.) Rouy. Bot. Mater. Gerb. Bot. Inst. Komarova Akad. Nauk S.S.S.R. 16: 425.
- Shahzadi I, Mehmood F, Ali Z, Ahmed I, Mirza, B, Abdullah . 2020. Chloroplast genome sequences of Artemisia maritima and Artemisia absinthium: comparative analyses, mutational hotspots in genus Artemisia and phylogeny in family Asteraceae. Genomics. 112(2):1454–1463.
- Song S, Zhu M, Wang Y, Huang W, He J, Zhang R. 2012. Essential oil useful for insecticide for killing homoptera insect i.e. cotton aphid and psyllid, obtained by steam distilling aerial part of Seriphidium borotalense. Chinese: CN113729036-A; CN113729036-B.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S., 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Vallès J, Garcia S, Hidalgo O, Martín J, Pellicer J, Sanz M, et al. 2011. Biology, Genome Evolution, Biotechnological Issues and Research Including Applied Perspectives in Artemisia (Asteraceae). In: Kader J, Delseny M, editors. Advances in Botanical Research Vol 60, vol. 60. London: Academic Press. p. 349–419.

