Abstract
The genus Cladosporium (Cladosporiaceae, Capnodiales) is a large genus of Ascomycota. Although the genus is mostly reported as saprobes from a wide range of substrates with a worldwide distribution, members of this genus comprise infectious agents in animals and plants. Of those, Cladosporium anthropophilum is a common saprophytic fungus and has been found to be a human opportunistic pathogen and plant pathogen. The complete mitochondrial genome of C. anthropophilum is characterized through the de novo assembly of Illumina sequencing data. The mitochondrial genome is a circular molecule of 35,937 bp with 30.23% GC content and has a total of 47 genes including 16 protein-coding genes, 29 transfer RNA genes, and two ribosomal RNA genes. Based on protein-coding sequences of the mitochondrial genome sequence, a phylogenetic tree was constructed to demonstrate the phylogenetic relationship of C. anthropophilum and its related genera.
The ascomycete genus Cladosporium (Cladosporiaceae, Capnodiales) introduced by Link (Link Citation1816) is mainly known as a ubiquitous environmental saprobic fungus with a worldwide distribution and isolated from a wide range of substrates (Bensch et al. Citation2012). In some cases, the members of this genus have been reported as etiologic agents in vertebrate hosts, including humans and animals (Sandoval-Denis et al. Citation2015) and plant endophytes and pathogens causing leaf spots on diverse herbaceous and woody plants (Bensch et al. Citation2012). Of those, Cladosporium anthropophilum Sandoval-Denis et al. (Citation2016) belongs to the C. cladosporioides species complex and is one of the most common saprophytic fungi given that it has been isolated quite frequently from a wide range of substrates including plant materials such as seeds or leaves (Tibpromma et al. Citation2019; Sandoval-Denis et al. Citation2016). The fungus can be further considered as a clinically relevant species given its prevalence from a set of clinical samples obtained from the USA (Sandoval-Denis et al. Citation2015).
An endophytic fungus, Cladosporium anthropophilum, was isolated from leaves of pecan from pecan orchards located in Miryang, South Korea (35°22′54.9″N 128°48′06.5″E). Of those successfully retained, the representative cultures were deposited to the culture collection (CDH) of the National Institute of Forest Science, South Korea (Accession No. CDH2021-02–05, nifos.forest.go.kr, Dong-Hyeon Lee, [email protected]) and a voucher specimen, CDH2021-03, was deposited to the Korean Agricultural Culture Collection (KACC), National Institute of Agricultural Sciences, South Korea (Accession No. KACC 49851, genebank.rda.go.kr). The DNA of the isolate CDH2021-03 was extracted from the fungal mycelium using CTAB DNA extraction method (Carter-House et al. Citation2020). Illumina paired-end (PE) library was constructed and sequenced using the Illumina HiSeqX platform with 151-bp PE reads. Raw sequencing data of 2.1 Gb were trimmed using the quality_trim program in CLC Assembly Cell package ver. 4.2.1 (QIAGEN, Denmark) with Phred scores > 20 and used for de novo assembly of mitochondrial genome according to the previous study (Lee et al. Citation2018, Cho et al. Citation2022). Trimmed high-quality read sequences of 1.8 Gb were de novo assembled using the clc_novo_assemble program with default parameters in the CLC Assembly Cell and then mitochondrial contigs were selected and ordered by similarity searches using BLAST against mitochondrial sequences from NCBI organelle genome resources (https://www.ncbi.nlm.nih.gov/genome/organelle/). The selected mitochondrial contigs were merged and gap-filled by a series of read mapping and extension to generate a complete and circularized mitochondrial genome. Sequence error was investigated and then corrected by read mapping of trimmed sequence data and manual curation. The complete mitogenome sequence was annotated using the GeSeq (Tillich et al. Citation2017) and Artemis (Carver et al. Citation2012) programs with reference mitochondrial genomes (GenBank accession nos. MN661341.1, MN657180.1, and MN657181.1). In addition, gene annotation was confirmed again by manual curation using BLAST searches against the reference mitochondrial genomes.
Mitochondrial genome of C. anthropophilum is a circular molecule of 35,937 bp with 30.23% GC content (GenBank accession number OK512878) and 1,640.6X mean coverage. A total of 47 genes were predicted in this mitochondrial genome, of which 16 are protein-coding genes including two free-standing open reading frames (ORFs), 29 transfer RNA genes, and two ribosomal RNA genes.
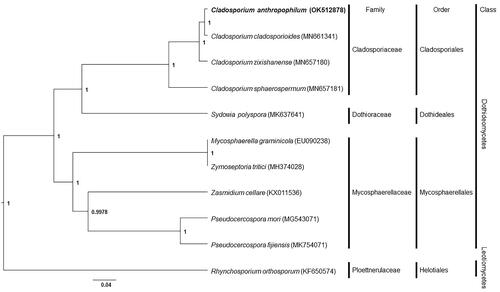
Phylogenetic analysis of C. anthropophilum with other taxa was performed using a Bayesian inference method with conserved 13 protein-coding sequences and revealed that C. anthropophilum is located in the same clade with other Cladosporium (). Among Cladosporium species in the phylogenetic tree, Cladosporium cladosporioides was the closest to C. anthropophilum. Mitochondrial genome (MN661341) of C. cladosporioides is 36,768 bp in length and has 46 genes (Liu et al. Citation2020). Compared to C. cladosporioides, C. anthropophilum mitochondrial genome in this study is 831-bp smaller and has a similar number of genes except for one additional tRNA gene for tRNA-Thr. In this study, the phylogenetic inference of the genus Cladosporium and its related taxa was demonstrated, and this would help understand species evolution and phylogenetic relationships between the related taxa.
Figure 1. Phylogenetic tree of mitochondrial genomes of Cladosporium anthropophilum and its related species. Thirteen protein-coding sequences conserved in the mitochondrial genomes of 11 species were multiple-aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html) and used to generate phylogenetic tree using Bayesian inference method of BEAST2 (Bouckaert et al. Citation2019) with default parameter. Posterior probability values are on the branches. The isolate, C. anthropophilum (KACC 49851), obtained in this study was represented by bold letters. GenBank accession nos. of mitochondrial genome sequences used for this tree are indicated within parentheses.
Author contributions
Dong-Hyeon Lee contributed to the conception of the study; Sung Eun Cho and Ji Yeon Oh performed the experiment and contributed significantly to analysis and manuscript preparation with constructive discussions; Dong-Hyeon Lee, Sung Eun Cho and Ji Yeon Oh wrote the manuscript and revised it critically for intellectual content. All authors approve the version to be published and agree to be accountable for all aspects of the work.
Regulation statement
This study did not require any ethical approval and specific permissions or licenses to perform the research.
Acknowledgments
The authors thank the owner of the pecan farm, Cheon-Wook Heo, who allowed us to collect samples on his farm, and Dr. Hyun-Oh Lee and Kyunghyun Nam in Phyzen Co. for their assistance withsoftware application and analyses.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OK512878. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA759657, SRR15693843, and SAMN21184963 respectively.
Additional information
Funding
References
- Bensch K, Braun U, Groenewald JZ, Crous PW. 2012. The genus cladosporium. Stud Mycol. 72:1–401.
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, et al. 2019. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 15(4):e1006650.
- Carter-House D, Stajich JE, Unruh S, Kurbessoian T. 2020. Fungal CTAB DNA Extraction. Protocols.io. doi:10.17504/protocols.io.bhx8j7rw
- Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 28(4):464–469.
- Cho SE, Oh JY, Lee DH. 2022. Complete mitochondrial genome sequence of Colletotrichum siamense isolated in South Korea. Microbiol Resour Announc. 11(5):e01055-21.
- Lee HO, Choi JW, Baek JH, Oh JH, Lee SC, Kim CK. 2018. Assembly of the mitochondrial genome in the Campanulaceae family using Illumina low-coverage sequencing. Genes. 9(8):383.
- Liu Y, Zhang G, Wang Y, Zhu K, Yang W, Wang Y, Yu H. 2020. Complete mitochondrial genome of Cladosporium cladosporioides YFCC 8621 isolated from a salt mine in Yunnan, southwestern China. Mitochondrial DNA Part B. 145(1):558–559.
- Sandoval-Denis M, Sutton DA, Martin-Vicente A, Cano-Lira JF, Wiederhold N, Guarro J, Gené J. 2015. Cladosporium species recovered from clinical samples in the United States. J Clin Microbiol. 53(9):2990–3000.
- Sandoval-Denis M, Gené J, Sutton DA, Wiederhold NP, Cano-Lira JF, Guarro J. 2016. New species of Cladosporium associated with human and animal infections. Pers: Mol Phylogeny Evol Fungi. 36(1):281–298.
- Tibpromma S, Mortimer P E, Karunarathna SC, Zhan F, Xu J, Promputtha I, Yan K. 2019. Morphology and multi-gene phylogeny reveal Pestalotiopsis pinicola sp. nov. and a new host record of Cladosporium anthropophilum from edible pine (Pinus armandii) seeds in Yunnan province, China. Pathogens. 8: 285.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Link HF. 1816. Observationes in ordines plantarum naturales. Observationes in ordines plantarum naturales. 2. Mag. Gesell. Naturf. Freunde Berlin. 8:25–45.
