Abstract
Phthiridium szechuanum is a bat surface parasite under the family Nycteribiidae that prefers to roost in the hair of bats to feed on their blood. In this study, the complete mitochondrial genome of P. szechuanum was studied for the first time using Illumina sequencing technology. The mitochondrial genome was 14,896 bp in size and was predicted to encode 37 genes including 13 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes. Phylogenetic trees were constructed using the IQ-TREE web server and phylogenetic analysis was performed using the maximum likelihood method, and P. szechuanum was found to be phylogenetically closest to Basilia ansifera. These data will provide a molecular biological approach to the species identification of P. szechuanum and provide a new reference for further studies on the population genetics and phylogeny of the family Nycteribiidae.
Introduction
The family Nycteribiidae is a class of obligate bat ectoparasites under the order Diptera and Hippoboscoidea, which together with the family Streblidae are known as bat flies (Porter et al. Citation2022). Members of the family Nycteribiidae have no wings, flat chests, small or missing eyes, and look like spiders. They like to live in caves inhabited by bats and feed on the blood of bats (Staegemann Citation2016). Previous studies have demonstrated that spider flies are important vectors for a variety of pathogens (Morse et al. Citation2012; Obame-Nkoghe et al. Citation2016). Nycteribiidae contains three subfamilies (Archinycteribiinae, Nycteribiinae, Cyclopodiinae), 11 genera, and 275 species, of which about 70% are distributed in the Western Hemisphere (Graciolli et al. Citation2007).
Phthiridium szechuanum (Theodor and Moscona Citation1954) is a species of the family Nycteribiidae, subfamily Nycteribiinae, genus Phthiridium. It is small (generally about 2 mm long) and usually lives on the fur on the back and abdomen of bats (Theodor Citation1967). At present, due to the lack of research on its genetic information, the phylogenetic research of P. szechuanum is not clear enough. In this study, we reported the complete mitogenome of P. szechuanum for the first time, this will help resolve outstanding questions related to the systematics and evolution of bat flies, and provide a better understanding of the phylogenetic relationships in the order of Diptera.
Materials and methods
Sample collection
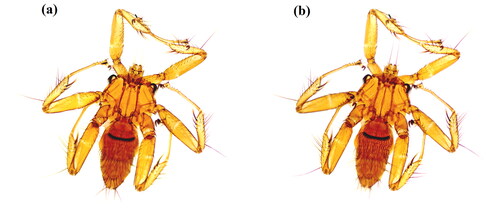
The specimens of P. szechuanum were collected in the Bat Cave (25°37′28ʺN, 103°33′54ʺE) in Zhanyi District, Qujing City, Yunnan Province. It was taken from the body surface of Rhinolophus sinicus, nine specimens were collected in total, and one specimen was selected as a voucher () and preserved in the Institute of Pathogens and Vector Biology of Dali University, Dali, China (https://www.dali.edu.cn/kxyj/yjs/1611.htm, voucher number: DLQJ2022001 Contact person: Xiaobin Huang, [email protected]), and the other eight were used for DNA extraction. Species identification was performed by Xianzheng Zhang based on morphological features using a Leica DM3000 LED biological microscope (Germany) according to the identification keys provided by Oskar Theodor (Theodor Citation1967). The sample was preserved in 95% ethanol and stored at −20 °C until utilization.
Figure 1. Due to the small size of the specimen, a stereomicroscope (Moticam 2506, China) was used to photograph P. szechuanum (♀, (a) is the back side, (b) is the ventral side).
Statement: All methods and procedures for capturing small mammals were performed by the guidelines and regulations approved by the Animal Ethics Committee of Dali University under approval number MECDU-202104-27.
DNA extraction, mitochondrial genome sequencing, and annotation
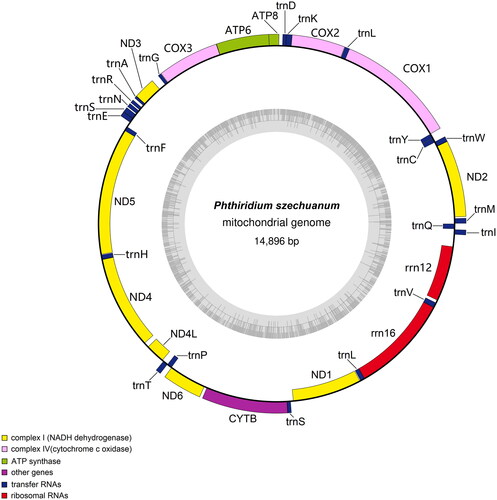
Firstly, the genomic DNA of P. szechuanum was extracted using DNeasy Blood and Tissue Kit (QIAGEN). Then, Sequencing of the complete mitochondrial genome on the Illumina Novoseq 6000 platform and used MitoZ 2.3 (https://doi.org/10.1093/nar/gkz173) to assemble the mitochondrial genome ((Meng et al. Citation2019). The complete mitochondrial genome sequence was annotated by MITOS and combined with GeSeq (Tillich et al. Citation2017) to predict the genomès coding genes, tRNA, and rRNA (Bernt et al. Citation2013). Finally, the mitochondrial genome cycle graph of P. szechuanum were mapped using OGDRAW (Greiner et al. Citation2019).
Sequence analysis
Geneious Prime was used to calculate the nucleobase composition of P. szechuanum. Carcinoscorpius rotundicauda JX437074 (Sarmiento et al. Citation2021) and Limulus Polyphemus JX983598 (Simpson et al. Citation2017) were used as outgroups, based on 13 mitochondrial protein-coding gene sequences, Mega X software (Kumar et al. Citation2018) was chosen to perform multiple sequence alignment of these sequences and a phylogenetic tree between P. szechuanum and 11 other species was constructed using the IQ-TREE web server (Trifinopoulos et al. Citation2016) using the maximum-likelihood method. Clade support was assessed using a nonparametric bootstrap method with 1000 replicates. The GenBank accession numbers of these 11 species are listed below. Sarcophaga albiceps KT444443 (Liao et al. Citation2016), Sarcophaga impatiens JN859549 (Nelson et al. Citation2012), Gasterophilus intestinalis KU236025 (Gao et al. Citation2016), Calliphora vicina JX913760 (Nelson et al. Citation2012), Calliphora vomitoria KT444440 (Ren et al. Citation2016), Ectophasia rotundiventris MK644821 (Li et al. Citation2017), Elodia flavipalpis JQ348961 (Zhao et al. Citation2013), Basilia ansifera MZ826150 (Porter et al. Citation2022), Dipseliopoda setosa MZ826151 (Porter et al. Citation2022), Paradyschiria parvula MK896865 (Trevisan et al. Citation2019), Paratrichobius longicrus MK896866 (Trevisan et al. Citation2019).
Results and discussion
Mitogenomic organization and nucleotide composition
The complete mitochondrial genome of P. szechuanum (GenBank accession number: OP459298) is 14,896 bp (), which contains 37 genes (13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes (12S and 16S), and one control region). The gene structure of P. szechuanum is similar to previous dipteran mitochondrial genome studies (Zhou et al. Citation2017). The nucleotide composition of P. szechuanum is 39.8% of A, 43.4% of T, 6% of G, and 10.8% of C. Among them, the proportion of A + T (83.2%) is higher than that of G + C (16.8%).
Protein-coding genes
Among the 13 PCGs, Five PCGs were started with ATG codon; atp6 and nad3 were started with ATA; atp8, nad2, nad6, and cytb were started with ATT; cox1 using TCG as a start codon, and nad5 was initialed by TTG codon. Six PCGs were terminated with TAA stop codon; while nad1, nad3, and cytb genes were stopped at TAG codon; cox2 and nad4 were ended with the incomplete stop codon TA; cox1 and nad5 were ended at single T.
Phylogenetic analysis
As shown in , within the Phylogenetic tree, the clustering results of each branch are consistent with those of the taxonomy. P. szechuanum was clustered together with species in the family Nycteribiidae and shared a close relationship with B. ansifera, supporting P. szechuanum among the family Nycteribiidae.
Conclusions
In this study, we reported the first complete mitochondrial genome sequencing of P. szechuanum. The mitochondrial genome sequence will be an important resource for further molecular studies with P. szechuanum, it can be used as a novel reference for further studies on the family Nycteribiidae.
Author contributions
Xianzheng Zhang, Xiaobin Huang, and Yujuan Wang planned and co-designed this research. Xianzheng Zhang performed the experiments, Huijuan Yang and Jinting Yang analyzed the data, Huijuan Yang and Xiaoyan Zheng constructed the phylogenetic tree. Xianzheng Zhang wrote the manuscript, Xiaobin Huang revised the manuscript. All authors have approved the manuscript for publication and agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession nos. OP459298. The associated Bio Project, BioSample, and SRA experiment numbers are PRJNA883607, SAMN30974214, and SRR21749916, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Gao DZ, Liu GH, Song HQ, Wang GL, Wang CR, Zhu XQ. 2016. The complete mitochondrial genome of Gasterophilus intestinalis, the first representative of the family Gasterophilidae. Parasitol Res. 115(7):2573–2579.
- Graciolli G, Autino AG, Claps GL. 2007. Catalogue of American Nycteribiidae (Diptera, Hippoboscoidea). Rev Bras Entomol. 51(2):142–159.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li X, Ding S, Hou P, Liu X, Zhang C, Yang D. 2017. Mitochondrial genome analysis of Ectophasia roundiventris (Diptera, Tachinidae). Mitochondrial DNA Part B. 2(2):457–458.
- Liao H, Yang X, Li Z, Ding Y, Guo Y. 2016. The complete mitochondria genome of Parasarcophaga albiceps (Diptera: sarcophagidae). Mitochondrial DNA Part A. 27(6):4696–4698.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Morse SF, Olival KJ, Kosoy M, Billeter S, Patterson BD, Dick CW, Dittmar K. 2012. Global distribution and genetic diversity of Bartonella in bat flies (Hippoboscoidea, Streblidae, Nycteribiidae). Infect Genet Evol. 12(8):1717–1723.
- Nelson LA, Lambkin CL, Batterham P, Wallman JF, Dowton M, Whiting MF, Yeates DK, Cameron SL. 2012. Beyond barcoding: a mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene. 511(2):131–142.
- Nelson LA, Cameron SL, Yeates DK. 2012. The complete mitochondrial genome of the flesh fly, Sarcophaga impatiens Walker (Diptera: Sarcophagidae). Mitochondrial DNA. 23(1):42–43.
- Obame-Nkoghe J, Rahola N, Bourgarel M, Yangari P, Prugnolle F, Maganga GD, Leroy E-M, Fontenille D, Ayala D, Paupy C. 2016. Bat flies (Diptera: Nycteribiidae and Streblidae) infesting cave-dwelling bats in Gabon: diversity, dynamics and potential role in Polychromophilus melanipherus transmission. Parasit Vectors. 9(1):1–12.
- Porter ML, Lutz H, Steck M, Chong RA. 2022. The complete mitochondrial genomes and phylogenetic analysis of two Nycteribiidae bat flies (Diptera: Hippoboscoidea). Mitochondrial DNA Part B. 7(8):1486–1488.
- Ren L, Guo Q, Yan W, Guo Y, Ding Y. 2016. The complete mitochondria genome of Calliphora vomitoria (Diptera: Calliphoridae). Mitochondrial DNA Part B. 1(1):378–379.
- Sarmiento ME, Chin KL, Lau NS, Aziah I, Ismail N, Norazmi MN, Acosta A, Yaacob NS. 2021. Comparative transcriptome profiling of horseshoe crab Tachypleus gigas hemocytes in response to lipopolysaccharides. Fish Shellfish Immunol. 117:148–156.
- Simpson SD, Ramsdell JS, Watson WH, III, Chabot CC. 2017. The draft genome and transcriptome of the Atlantic horseshoe crab, Limulus polyphemus. Int J Genomics. 2017:1–14.
- Staegemann MW. 2016. [Drivers of bat fly diversity and prevalence of six Rhinolophus bat species in southern Africa] [doctoral dissertation]. Westville: University of KwaZulu-Natal.
- Theodor O, Moscona A. 1954. On the bat parasites in Palestine I. Nycteribiidae, Streblidae, Hemiptera, Siphonaptera. Parasitology. 44(1-2):157–245.
- Theodor O. 1967. An illustrated catalogue of the Rothschild collection of Nycteribiidae (Diptera) in the British Museum (Natural History); with keys and short descriptions for the identification of subfamilies, genera, species and subspecies. London: British Museum (Natural History).
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Trevisan B, Alcantara DM, Machado DJ, Marques FP, Lahr DJ. 2019. Genome skimming is a low-cost and robust strategy to assemble complete mitochondrial genomes from ethanol preserved specimens in biodiversity studies. PeerJ. 7:e7543.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Zhao Z, Su TJ, Chesters D, Wang SD, Ho SYW, Zhu CD, Chen XL, Zhang CT. 2013. The mitochondrial genome of Elodia flavipalpis Aldrich (Diptera: Tachinidae) and the evolutionary timescale of tachinid flies. PLOS One. 8(4):e61814.
- Zhou Q, Ding S, Li X, Zhang T, Yang D. 2017. Complete mitochondrial genome of Allognosta vagans (Diptera, Stratiomyidae). Mitochondrial DNA Part B. 2(2):461–462.