Abstract
Paratanakia chii is a bitterling fish of the genus Paratanakia, subfamily Acheilognathinae and family Cyprinidae. The mitochondrial DNA sequence of P. chii is reported in this paper. The complete mitochondrial genome of P. chii is 16,575 bp in length, including 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and 1 displacement loop (D-loop). The genome sequence is consistent with those of most other carp. The majority of PCGs have AT- (Met) start codons and TA- end codons. The A + T contents of the genome, PCGs, transfer RNAs (tRNAs), and ribosomal RNAs (rRNAs) are 56.92%, 58.07%, 56.34%, and 54.21%, respectively. Phylogenetic analysis showed that P. chii is most closely related to Tanankia himantegus. These data will benefit relative ecological and phylogenetic studies.
The rainbow bitterling, Paratanakia chii Miao, 1934 (Cypriniformes: Cyprinidae), inhabits slow-moving or stationary waters and is widely distributed in East Asia, Southeast Asia, and Europe. (Xiong et al. Citation2017). This fish is small, colorful, and diverse in shape, has high ornamental value and is increasingly loved by people (Chang et al. Citation2014). Thus, overfishing, water pollution, and damming are likely impacting the specie. The majority of studies on this species are focused on the differentiation of visual spectra and nuptial colorations in response to the varied photic circumstances of their habitats, but little information on mitochondrial sequences is available (Chang et al. Citation2015). We sequenced the whole mitogenome of P. chii in this work to provide information for the identification of improved markers that can be applied in the classification of this family as well as a helpful reference for the conservation of the species ().
A specimen of P. chii was collected in May 2022 from the Rongjiang River of Jieyang County, Guangdong, China (23.56°N, 116.42°E), and then the live fish was stored in an oxygenated container. Genomic DNA was extracted from the fins. A specimen was deposited at the College of Fisheries, Zhejiang Ocean University (contact: Haixia Zhang, [email protected]) under the voucher number ZHX-202206-15. The phylogenetic tree was confirmed using the maximum likelihood method in MEGA 7 using a bootstrap consensus tree derived from 1000 replicates; no significant changes in tree topologies were noted (Krzyżanowska et al. Citation2019).
DNA was extracted from the tissue samples by using the Tissue Cell Genome DNA Rapid Extraction Kit (Beijing Adelaide Biotechnology Co., Ltd.) and then subjected to gel electrophoresis to assess its quality before the subsequent experiments. The PCR products were sequenced using an ABI 3730 automatic sequencer (Sanger sequencing) and further assembled manually using DNAStar v7.1 software. The mt genome was roughly annotated following the procedure described previously. First, raw mitogenomic sequences were loaded into MITOS web servers to determine gene boundaries. Searching for ORFs yielded the precise sites of protein-coding genes (PCGs) (employing genetic code 2, the vertebrate mitochondrion). ARWEN, DOGMA, and MITOS were used to identify all tRNAs (Zhang et al. Citation2017). The precise boundaries of rrnL and rrnS were determined via a comparison with homologs.
We constructed a phylogenetic tree including P. chii and 14 other fishes from the GenBank database. For each species, we extracted the 13 PCGs and concatenated them into one sequence (Thierry-Mieg and Thierry-Mieg Citation2006) (). The topology of the tree was inferred using the maximum likelihood method in the program MEGA 7 (ML, based on the GTR + G + I model) with 1000 bootstrap replicates (Meier et al. Citation2017). The species used for tree construction were as follows: Tanakia limbata (Luo et al. Citation2016), Tanakia lanceolata (Xu et al. Citation2016), Tanakia himantegus (Cheng and Tao Citation2016), Rhodeus lighti (Thierry-Mieg and Thierry-Mieg Citation2006), Rhodeus sinensis (Zhu et al. Citation2016), Rhodeus sericeus (Xu et al. Citation2016), Rhodeus uyekii (Kim et al. Citation2006), Rhodeus shitaiensis (Li et al. Citation2015), Rhodeus fangi (Yu et al. Citation2016), Rhodeus suigensis (Hwang et al. Citation2014), Acheilognathus barbatus (Tao and Sun Citation2016), Acheilognathus tabira erythropterus (Nagata and Kitamura Citation2019), Acheilognathus omeiensis (Zou et al. Citation2018), Acheilognathus hypselonotus (Zhu et al. Citation2021), Rhodeus ocellatus (Hu et al. Citation2016), Acheilognathus tonkinensis (Zhang et al. Citation2018), and Silurus asotus (Yang et al. Citation2019).
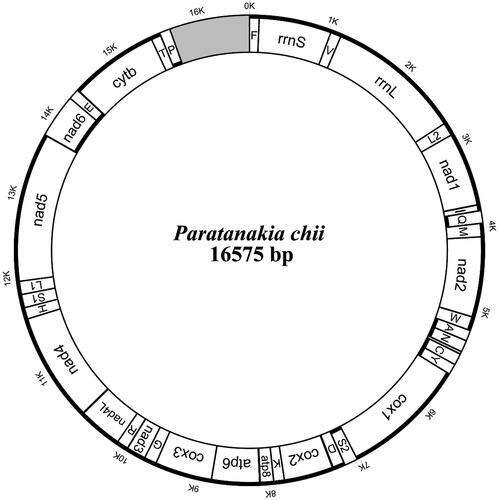
The complete mitochondrial genome of P. chii is 16,575 bp in length and has the typical structure observed in vertebrates (GenBank accession number: ON961028). The genome contains 38 sequence elements, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and a noncoding control region (D-loop region). The total base composition of the mitogenome is A (29.88%), T (27.04%), C (26.55%), and G (16.53%). The A + T contents of the genome, PCGs, transfer RNAs (tRNAs), and ribosomal RNAs (rRNAs) are 56.92%, 58.07%, 56.34%, and 54.21%, respectively. For the 13 protein-coding genes, 12 genes start with ATG, while only COI initiates with the GTG codon (Arca et al. Citation2012). For each of the 15 mitogenomes, there are two different starting codon types (ATG and GTG) and three different stop codon (Ma et al. Citation2022) types (TAA, TAG, and T-). Our results show that among the 13 PCGs, ND5 is the longest gene (1836 bp), and ATP8 is the shortest gene (165 bp). The lengths of the 12S and 16S ribosomal RNA genes are 960 bp and 1684 bp, respectively, which are located between tRNA-Phe and tRNA-Leu and separated by the tRNA-Val gene. All 22 typical tRNAs possess a complete secondary structure, ranging from 69 bp to 76 bp. To determine the phylogenetic relationships of both species in the Cyprinidae family, we used MEGA7 software to construct a phylogenetic tree including P. chii and 14 other fishes based on maximum likelihood (ML). The phylogenetic position of P. chii was shown. Phylogenetic analysis supported the close genetic relationship between P. chii and Tanankia chii, indicating that the two species share a more recent common ancestor gene (). The GenBank accession numbers of 15 fishes are given in parentheses.
Figure 3. The phylogenetic tree is based on the complete mitochondrial genomes of Paratanakia chii and other 14 species.
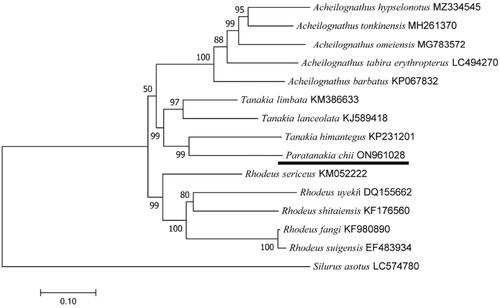
Our study describes the mitochondrial genome and phylogenetic relationships of P. chii. The results of this study indicate that P. chii and Tanankia. himantegus cluster into a single clade; therefore, we infer that these two specimens of Paratanakia and Tanakia are most likely the same species. These data will be useful for further molecular identification and population genetic studies of this species.
Ethical approval
Experiments were performed in accordance with the recommendations of the Ethics Committee of Zhejiang Ocean University. The sample of this study did not involve endangered or protected animals, and the experiment followed the Laboratory animal—Guideline for ethical review of animal welfare of the National Standard of the People’s Republic of China (GB/T 35892-2018).
Author contributions
Haixia Zhang and Zhangjie Chu designed the experiments and wrote the original manuscripts; Dan Zhao, Chang Hu, Wenli Duan, and Hang Chu revised the text; Zhangjie Chu and Dan Zhao collected samples; and Haixia Zhang, Wenli Duan, Hang Chu, Dan Zhao, and Hang Chu analyzed the data. All authors gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (24.3 KB)Supplemental Material
Download MS Word (10.8 KB)Supplemental Material
Download PDF (77.4 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data obtained in this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number ON961028.
Additional information
Funding
References
- Arca M, Hinsinger DD, Cruaud C, Tillier A, Bousquet J, Frascaria-Lacoste N. 2012. Deciduous trees and the application of universal DNA barcodes: a case study on the circumpolar Fraxinus. PLOS One. 7(3):e34089.
- Chang CH, Li F, Shao KT, Lin YS, Morosawa T, Kim S, Koo H, Kim W, Lee JS, He S, et al. 2014. Phylogenetic relationships of Acheilognathidae (Cypriniformes: Cyprinoidea) as revealed from evidence of both nuclear and mitochondrial gene sequence variation: evidence for necessary taxonomic revision in the family and the identification of cryptic species. Mol Phylogenet Evol. 81:182–194.
- Chang CH, Shao YT, Fu WC, Anraku K, Lin YS, Yan HY. 2015. Differentiation of visual spectra and nuptial colorations of two Paratanakia himantegus subspecies (Cyprinoidea: Acheilognathidae) in response to the distinct photic conditions of their habitats. Zool Stud. 54(1):e41.
- Cheng Y, Tao W. 2016. The complete mitogenome of Tanakia himantegus (Cypriniformes; Cyprinidae). Mitochondrial DNA Part A. 27(6):4144–4145.
- Hu J, Chen Y, Zhao H, Yang H, Liu L. 2016. Complete mitochondrial genome of Rhodeus ocellatus (Cypriniformes: Cyprinidae). Mitochondrial DNA Part A. 27(5):3489–3490.
- Hwang DS, Lee WO, Lee JS. 2014. Complete mitochondrial genome of the freshwater fish, Rhodeus suigensis (Cypriniformes, Cyprinidae). Mitochondrial DNA. 25(1):5–6.
- Kim BC, Kang TW, Kim MS, Kim CB. 2006. The complete mitogenome of Rhodeus uyekii (Cypriniformes, Cyprinidae). DNA Seq. 17(3):181–186.
- Krzyżanowska DM, Maciąg T, Ossowicki A, Rajewska M, Kaczyński Z, Czerwicka M, Rąbalski Ł, Czaplewska P, Jafra S. 2019. Ochrobactrum quorumnocens sp. nov., a quorum quenching bacterium from the potato rhizosphere, and comparative genome analysis with related type strains. PLOS One. 14(1):e0210874.
- Li F, Shao KT, Lin YS, Chang CH. 2015. The complete mitochondrial genome of the Rhodeus shitaiensis (Teleostei, Cypriniformes, Acheilognathidae). Mitochondrial DNA. 26(2):301–302.
- Luo Y, Cao X, Zhu Y. 2016. The complete mitochondrial genome of Tanakia limbata (Cypriniformes: Cyprinidae). Mitochondrial DNA Part A. 27(3):1713–1714.
- Ma L, Xing J, Li Q, Zhang Z, Xu K. 2022. Development of a universal antibiotic resistance screening reporter for improving efficiency of cytosine and adenine base editing. J Biol Chem. 298(7):102103.
- Meier JI, Marques DA, Mwaiko S, Wagner CE, Excoffier L, Seehausen O. 2017. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat Commun. 8:14363.
- Nagata N, Kitamura JI. 2019. The complete mitochondrial genomes of two endangered bitterling Acheilognathus tabira tohokuensis and A. tabira erythropterus (Cyprinidae, Acheilognathinae). Mitochondrial DNA Part B 4(2):2865–2866.
- Tao W, Sun L. 2016. The complete mitogenome of Acheilognathus barbatus (Cypriniformes; Cyprinidae). Mitochondrial DNA Part A. 27(3):2274–2275.
- Thierry-Mieg D, Thierry-Mieg J. 2006. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 7(Suppl 1):S12.
- Xiong LW, Wang Q, Wang SB, Wang JG, Feng Q, Yue LJ. 2017. Description of 21 microsatellites for the Chinese bitterling, Rhodeus sinensis Günther, 1868. J Appl Ichthyol. 33(5):940–942.
- Xu C, Xie F, Zhang X, Zhao S. 2016. The complete mitochondrial genome of Amur bitterling (Rhodeus sericeus) from China. Mitochondrial DNA Part A. 27(4):2377–2378.
- Xu X, Cao X, Zhu Y. 2016. The complete mitochondrial genome of Tanakia lanceolata (Cypriniformes: Cyprinidae). Mitochondrial DNA Part A. 27(2):867–868.
- Yang N, Li Y, Liu Z, Chen Q, Shen Y. 2019. The complete mitochondrial genome of Silurus asotus (Siluriformes: Siluridae: Silurus) and its phylogenetic analysis. Mitochondrial DNA Part B. 4(2):2377–2378.
- Yu Y, Yi W, Ma Z, Yang R, Shen J. 2016. The complete mitochondrial genome of Rhodeus fangi (Cypriniformes, Cyprinidae, Acheilognathinae). Mitochondrial DNA Part A. 27(1):284–285.
- Zhang D, Zou H, Wu SG, Li M, Jakovlić I, Zhang J, Chen R, Wang GT, Li WX. 2017. Sequencing of the complete mitochondrial genome of a fish-parasitic flatworm Paratetraonchoides inermis (Platyhelminthes: Monogenea): tRNA gene arrangement reshuffling and implications for phylogeny. Parasit Vectors. 10(1):462.
- Zhang M, Wang K, Zhang L, Tang Z, Zhong S, Tan H, Wu L, Chen X, Cheng G. 2018. Complete mitochondrial genome and phylogenetic implications of the rainbow bitterling Acheilognathus tonkinensis (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B. 3(2):1178–1179.
- Zhu X, Ma Z, Yang X, Xu H, Yang R. 2016. Complete mitochondrial genome of the Chinese bitterling Acheilognathus macropterus (Cypriniformes: cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):589–590.
- Zhu L, Che X, Liu X, Wang X, Wang J, Cheng G, Chen X, Chen X. 2021. Complete mitochondrial genome of Acheilognathus hypselonotus Bleekers (Cypriniformes: Cyprinidae) in China’s Dianshan Lake. Mitochondrial DNA Part B. 6(12):3367–3368.
- Zou Y, Chen M, Wu T, Tian T, Wen Z. 2018. Identification and analysis of the complete mitochondrial genome of Acheilognathus omeiensis (Cypriniformes, Cyprinidae). Mitochondrial DNA B. 3(1):270–271.