Abstract
Salvia chienii E.Peter is a medicinal herb mainly distributed in Huangshan Mountain of Anhui province, China. In this study, the first complete chloroplast genome of S. chienii was sequenced and assembled. The genome length was 151,530 bp and encoded 143 genes (91 protein-coding genes, eight rRNA genes, and 37 tRNA genes). The phylogenomic analysis showed that S. chienii was closely related to S. miltiorrhiza. Further evolutionary studies of the genus Salvia could benefit from the complete chloroplast genome of S. chienii present in this study.
Introduction
The genus Salvia is the largest member of the Lamiaceae family and is widely distributed around the world. Some Salvia species have antioxidant, anti-inflammatory and anti-cancer properties (Bonesi et al. Citation2017). Their main active constituents are phenolic acids and terpenoids. Salvia chienii E.Peter is primarily found in Huangshan Mountain of Anhui Province, China. The high content of salvianolic acid B and low levels of tanshinones were recorded in this species. In the ‘Flora of China’ (Wu and Raven Citation1999), it was mentioned that S. chienii generally grows along roadsides or hillsides and is a perennial plant with upright stems covered with profuse pubescence from the top downward. It has trifoliate or simple leaves and usually blooms in July. Chloroplast genomes of numerous Salvia species have been published (Zhou et al. Citation2020, Hu et al. Citation2020), but the chloroplast genome of S. chienii is not reported yet. In the study, we reported the complete chloroplast genome of S. chienii for the first time, which helped to clarify interspecific relationships among the Salvia species.
Materials and methods
Fresh samples of S. chienii were collected from Huangshan, Anhui, China (GPS: E114°04′03.15″, N31°49′46.46″) (). A specimen was deposited at Herbarium of Shanghai Chenshan Botanical Garden (CSH) (Yukun Wei, [email protected]) and was given a voucher number S1094. Total genomic DNA was extracted by using DNA Plantzol reagent (Invitrogen, Carlsbad, CA, USA). A 350-bp paired-end library was constructed using Illumina’s TruSeq Nano DNA Library Preparation kit following the manufacturer’s instructions. About 3.51 G of original data was sequenced using Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA). The chloroplast genome was assembled using rbcL gene (genbank number: NC_023209) as a seed sequence by NOVOPlasty (Dierckxsens et al. Citation2017), and annotated with GeSeq v2.03 and Geneious Prime 2022 (https://www.geneious.com). The newly annotated chloroplast genome was submitted to GenBank with accession number OK094518.
Figure 1. Image of S. chienii species. Yukun Wei collected the image in Tangkou Town, Huangshan, Anhui, China (GPS: E114°04′03.15″,N31°49′46.46″). The leaves of S. chienii are long oval with serrated edges. Verticillasters form extended racemes with lavender flowers.

To reveal the phylogenetic relationship of S. chienii, the sequences of other 23 chloroplast genomes were downloaded from NCBI (https://www.ncbi.nlm.nih.gov). Callicarpa nudiflora was used as an outgroup. Alignments were performed by MAFFT v7.486 (Katoh and Standley Citation2013), and a maximum likelihood tree was constructed using IQ-TREE (Nguyen et al. Citation2015) with the best-predicted model TVM + F+R3 and 1000 bootstrap replicates. Bayesian reference were conducted by Mrbayes (Ronquist et al. Citation2012) (v 3.2.7).
Results
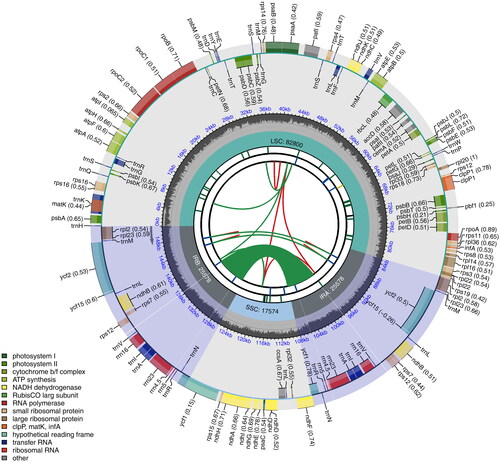
The size of the chloroplast genome of S. chienii was 151,530 bp with a typical circular structure and a GC content of 38% (). It consisted of a large single copy region (LSC) of 82,800 bp, a small single copy region (SSC) of 17,574 bp, and two inverted repeating regions (IR) of 25,578 bp. A total of 143 genes were annotated in the chloroplast genome of S. chienii, including 91 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Among these genes, seven protein-coding (ndhB, rpl2, rpl23, rps12, rps7, ycf15 and ycf2), seven tRNA (trnA-UgC, trnI-Cau, trnI-Gau, trnL-caa, trnN-guu, trnR-ACG and trnV-GAC) and four rRNA genes (rrn5, rrn4.5, rrn23, rrn16) have two copies. Totally 11 cis-splicing genes including rps16, atpF, rpoC1, pafl, clpP1, petB, petD, rpl16, rpl2, ndhB and ndhA (Figure S2), and one trans-splicing genes rps12 (Figure S3) were detected by CPGview (Liu et al. Citation2023) (http://www.1kmpg.cn/cpgview/).
Figure 2. The chloroplast genome map of S. chienii chlobroplast genome. The length of genome is presented in the inner circle. Large single copy (LSC), Single short copy (SSC) and inverted repeat regions (IRA and IRB) are marked. A total of 143 annotated genes are presented in the outer circle, consisting of 91 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Genes were classified by their function in colors. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and Palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. Black: c (complex repeat); Green: p1 (repeat unit size = 1); Yellow: p2 (repeat unit size = 2); Purple: p3 (repeat unit size = 3); Blue: p4 (repeat unit size = 4); Orange: p5 (repeat unit size = 5); Red: p6 (repeat unit size = 6). The small single-copy (SSC), inverted repeat (IRA and IRB), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. Genes are color-coded by their functional classification. The functional classification of the genes is shown in the bottom left corner.
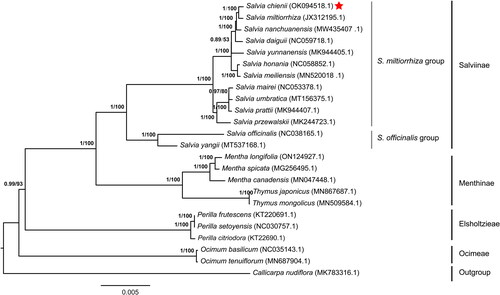
IQTree conducted the phylogenetic analysis with 24 chloroplast genomes (). As shown in the figure, six clades were generated in the phylogenetic tree, consisting of Salviinae, Menthinae, Elsholtzieae, Ocimeae and outgroup. Our results showed that Salviinae had two subgroups (S. miltiorrhiza subgroup and S. officinalis subgroup). The separation time of S. officinalis subgroup was earlier than S. miltiorrhiza subgroup. The clustering results supported that S. chienii was more closely related to S. miltiorrhiza, as the two species were clustered together under S. miltiorrhiza subgroup.
Figure 3. Phylogeny of Lamiaceae based on complete chloroplast genomes, accession numbers were listed behind each taxon. Statistical support values were shown on nodes. The complete chloroplast genomes used for constructing the phylogenetic tree contains S. miltiorrhiza (Hu et al. Citation2020) (JX312195.1), Callicarpa nudiflora (Wang et al. Citation2019) (MK783316.1), Mentha canadensis (Huaizhu et al. Citation2019) (MN047448.1), Mentha longifolia (Zubair Filimban et al. Citation2022)(ON124927.1), Mentha spicata (Wang et al. Citation2017) (MG256495.1), Ocimum basilicum (Rabah et al. Citation2017) (NC035143.1), Ocimum tenuiflorum (Kavya et al. Citation2021) (MN687904.1), Perilla citriodora (Mo et al. Citation2017) (KT22690.1), Perilla frutescens (Shen et al. Citation2016) (KT220691.1), Perilla setoyensis (NC030757.1; Unpublished), Salvia daiguii (Zhou et al. Citation2020) (NC059718.1), Salvia honania (Wang et al. Citation2022) (NC058852.1), Salvia mairei (NC053378.1; Unpublished), Salvia meiliensis (Su et al. Citation2022) (MN520018.1), Salvia nanchuanensis (Su et al. Citation2022) (MW435407.1), S. officinalis (Du et al. Citation2022) (NC038165.1), Salvia prattii (MK944407.1; Unpublished), Salvia przewalskii (Du et al. Citation2019) (MK344723.1), Salvia umbratica (MT156375.1; Unpublished), Salvia yangii (Cao et al. Citation2020) (MT537168.1), Salvia yunnanensis (Tao et al. Citation2019) (MK944405.1), Thymus japonicus (Kim et al. Citation2020) (MN867687.1), Thymus mongolicus (Huaizhu et al. Citation2020) (MN509584.1). The type of sequences used for building the phylogenetic tree are the complete chloroplast genomes downloaded from NCBI (https://www.ncbi.nlm.nih.gov/), and built with IQTree with the best predicted model TVM + F+R3 and 1000 bootstrap replicates. Bayesian posterior probabilities and bootstrap scores were present on the tree nodes. Bayesian posterior probabilitis were culculated by Mrbayes (v3.2.7). S. chienii is close to S. miltiorrhiza.
Discussion
Salvia species belongs to multi-lineage evolution, but the relationship between the lineages is still unclear. The evolutionary relationships of Salvia species are frequently investigated using phylogenetic analysis. Our phylogenetic tree showed that there are two subgroups of Salvia species; S. miltiorrhiza subgroup which comprises Salvia species in East Asia and S. officianlis subgroup which comprises Salvia species in Europe. The divergence between the two subgroups is consistent with what was previously reported (Hu et al. Citation2018). S. miltiorrhiza group has two core subgroups, which suggests the evolutionary relationships of Salvia species in Eastern Asia. However, relationships within S. miltiorrhiza group remain unclear (Hu et al. Citation2018). S. chienii is clustered with S. miltiorrhiza, which will provide valuable information to explore the lineage of Salvia species in Eastern Asia.
Ethical approval
Research and collection of plant material were conducted according to the guidelines provided by Zhejiang Sci-Tech University. The plant was collected with special permission from Shanghai Chenshan Botanical Garden. This article does not contain any studies with endangered or protected species performed by any of the authors. Experiments were performed following the recommendations of the Ethics Committee of Zhejiang Sci-Tech University.
Authors’ contributions
The experiments were conceived and organized by Dongfeng Yang and Yukun Wei. Ying Cheng was involved in the design, data analysis, and paper drafting. Fuhai Yuan and Yukun Wei contributed to the collection of plant material. The paper was written by Ying Cheng and Dongfeng Yang, and revised by Fuhai Yuan and Yelong Sheng, Dongfeng Yang, Ann Abozeid and Yanbo Huang. All authors agree to be accountable for all aspects of the final manuscript.
Supplemental Material
Download JPEG Image (194.7 KB)Supplemental Material
Download JPEG Image (409.6 KB)Supplemental Material
Download JPEG Image (717.7 KB)Disclosure statement
The authors report no conflicts of interest. Fuhai Yuan and Yelong Sheng are the employee of Changzhou Menghe Shuangfeng Chinese Herbal Medicine Technology, and they provided the funding of Science and Technology Support Program (Agriculture) of Changzhou Jiangsu Province (CE20212032) for the study. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data supporting this study’s findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under accession no. OK094518. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA763313, SRR15901094, and SAMN21437815, respectively.
Additional information
Funding
References
- Bonesi M, Loizzo MR, Acquaviva R, Malfa GA, Aiello F, Tundis R. 2017. Anti-inflammatory and antioxidant agents from Salvia Genus (Lamiaceae): an assessment of the current state of knowledge. Antiinflamm Antiallergy Agents Med Chem. 16(2):70–86.
- Cao M-T, Wu J-J, Wang R-H, Xu L, Qi Z-C, Wei Y-K. 2020. The complete chloroplast genome of Russian sage Salvia yangii B. T. Drew (Lamiaceae). Mitochondrial DNA Part B Resour. 5(3):2590–2591.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Du Q, Yang H, Zeng J, Chen Z, Zhou J, Sun S, Wang B, Liu C. 2022. Comparative genomics and phylogenetic analysis of the chloroplast genomes in three medicinal Salvia species for bioexploration. IJMS. 23(20):12080.
- Du Y, Wang Y-Y, Xiang C-L, Yang M-Q. 2019. Characterization of the complete chloroplast genome of Salvia Przewalskii Maxim. (Lamiaceae), a substitute for Dan-Shen Salvia miltiorrhiza Bunge. Mitochondrial DNA Part B Resour. 4(1):981–982.
- Hu G-X, Takano A, Drew BT, Liu E-D, Soltis DE, Soltis PS, Peng H, Xiang C-L. 2018. Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Ann Bot. 122(4):649–668.
- Hu J, Zhao M, Hou Z, Shang J. 2020. The complete chloroplast genome sequence of Salvia miltiorrhiza, a medicinal plant for preventing and treating vascular dementia. Mitochondrial DNA Part B Resour. 5(3):2460–2462.
- Huaizhu L, Bai L, Bai J, Wang P, Zhou C, Lingling D, Jiang J, Liu J, Wang Q. 2020. The complete chloroplast genome sequence of Thymus mongolicus (Labiatae), a special spice plant. Mitochondrial DNA Part B Resour. 5(3):2597–2598.
- Huaizhu L, Lin Y, Bai J, Qiuyan A, Lingling D, Rui-Xue S. 2019. The complete chloroplast genome sequence of Mentha canadensis (Labiatae), a traditional Chinese herbal medicine. Mitochondrial DNA Part B Resour. 5(1):55–56.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kavya NM, Balaji R, Parani M, Senthilkumar, P, Tanuja . 2021. The complete chloroplast genome of Ocimum tenuiflorum L. subtype Krishna Tulsi and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 6(8):2358–2360.
- Kim HT, Pak JH, Kim JS. 2020. The complete chloroplast genome sequence of Thymus quinquecostatus var. japonicus (Lamiaceae), an endemic to Ullenung Island of Korea. Mitochondrial DNA Part B Resour. 5(3):2401–2402.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: A package for visualizing detailed chloroplast genome structures. Molecular Ecology Resources. 1–11.
- Mo J-S, Kim K, Lee MH, Lee J-H, Yoon U-H, Kim T-H. 2017. The complete chloroplast genome sequence of Perilla citriodora (Makino) Nakai. Mitochondrial DNA Part A DNA Mapp Seq Anal. 28(1):131–132.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Rabah SO, Lee C, Hajrah NH, Makki RM, Alharby HF, Alhebshi AM, Sabir JS, Jansen RK, Ruhlman TA. 2017. Plastome sequencing of ten nonmodel crop species uncovers a large insertion of mitochondrial DNA in Cashew. The Plant Genome. 10(3).
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shen Q, Yang J, Lu C, Wang B, Song C. 2016. The complete chloroplast genome sequence of Perilla frutescens (L.). Mitochondrial DNA Part A. 27:3306–3307. doi: 10.3109/19401736.2015.1015015.
- Su T, Geng Y-F, Xiang C-L, Zhao F, Wang M, Gu L, Hu G-X. 2022. Chloroplast genome of Salvia Sect. Drymosphace: comparative and phylogenetic analysis. Diversity Accepted. 14(5):324.
- Tao A, Zhao F, Qian J. 2019. The complete chloroplast genome sequence of the medicinal plant Salvia yunnanensis C. H. Wright. (Lamiaceae). Mitochondrial DNA Part B Resour. 4(2):3603–3605.
- Wang HX, Chen L, Cheng XL, Chen WS, Li LM. 2019. Complete plastome sequence of Callicarpa nudiflora Vahl (Verbenaceae): a medicinal plant. Mitochondrial DNA Part B Resour. 4(2):2090–2091.
- Wang K, Li L, Hua Y, Zhao M, Li S, Sun H, Lv Y, Wang Y. 2017. The complete chloroplast genome of Mentha spicata, an endangered species native to south Europe. Mitochondrial DNA Part B Resour. 2(2):907–909.
- Wang Z-X, Huang Y-B, Yuan F-H, Lin J-J, Xiao H-W, Yang D-F, Wei Y-K. 2022. Characterization of complete chloroplast genome of Salvia honania L. H. Bailey (Lamiaceae). Mitochondrial DNA Part B Resour. 7(8):1474–1476.
- Wu Z, Raven P. 1999. Flora of China. Vol. 17. Beijing(China): Science Press.
- Zhou X, Huang Y-B, Zhang Z-C, Xu X-Y, Wang R-H, Xu L, Qi Z-C, Wei Y-K. 2020. The complete chloroplast genome of endangered Zhangjiajie sage Salvia daiguii Y. K. Wei & Y. B. Huang (Lamiaceae). Mitochondrial DNA. Part B Resour. 5(4):3833–3834.
- Zubair Filimban F, Yarádua SS, Bello A, Choudhry H. 2022. Complete chloroplast genomes of two mint species (Lamiaceae) from Al-Madinah, Saudi Arabia: phylogenetic and genomic comparative analyses. Mitochondrial DNA Part B Resour. 7(10):1797–1799.
