Abstract
Gortyna guizhouensis Wu, Yang, and Han, 2022 (Lepidoptera, Noctuidae) is a newly discovered moth species that feeds on tobacco piths during the larval stage. In this study, we sequenced and analyzed the complete mitochondrial genome (mitogenome) of G. guizhouensis larvae via next-generation sequencing. The mitogenome was 15,441 bp long with an overall A + T content of 79.1%, 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA subunit genes and one control region. The phylogenetic tree, based on the nucleic acid sequences of 13 shared PCGs of 20 Noctuidae species, revealed that G. guizhouensis is in a well-supported clade with Striacosta albicosta.
Introduction
Tobacco is an important cash crop that, until recently, was host to only one stem borer pest species, Scrobipalpa heliopa Loew 1900, in China. However, in 2020, a new tobacco stem borer pest was discovered in Guizhou province. It has been identified as a new species of the Gortyna genus in the Noctuidae family (Noctuoidea superfamily), and has been named Gortyna guizhouensis Wu, Yang, & Han, Citation2022 (Lepidoptera, Noctuidae) (Wu et al. Citation2022) (). The Gortyna genus is widely distributed throughout South Asia, the Middle East, Europe, and North Africa (Gardiner et al. Citation2017). G. guizhouensis is a new species of which no genomic sequencese were available so far, thus mitochondrial genomic data will be conducive to DNA classification and identification of such pests in the future.
Figure 1. Gortyna guizhouensis Wu, Yang, and Han, Citation2022 (Lepidoptera, Noctuidae): (A) Larva. The larva is black and orange-yellow; (B) Adult. The ground color of the forewings is golden-red and veins are indistinct; a dark streak is present between the orbicular and reniform spots. Photos by Meng Leng.

Materials
In this study, we sequenced the complete mitochondrial genome of G. guizhouensis. The larvae were collected in Weining County (104°7′57′′E, 27°15′45′′N), Guizhou Province, China in June 2021. Specimens were deposited at the Institute of Entomology, Guizhou University, Guiyang, China (Hai-yan Zhao, [email protected]) under the voucher number Gzh-2021-017.
Methods
We used the DNeasy Blood and Tissue Kit (cat.nos.69504 and 69506, QIAGEN, Hilden, Germany) to extract total DNA from the larvae head. An Illumina ReSeq library was prepared, with an average insert size of 400 bp. The library was sequenced paired-end sequenced (2 × 150 bp) using the Illumina NovaSeq6000 platform (Berry Genomics, Beijing, China). We assembled and annotated the complete mitogenome sequence using NOVOPlasty v2.7.2 (Dierckxsens et al. Citation2017), with default settings for the K-mer value, and MitoZ v2.4-alpha (Meng et al. Citation2019) was used to annotate the genome. Samtools v1.1 (Li et al. Citation2009) was used to assess sequencing depth based on BAM files, and circlize v0.4.16 was used to plot circles.
Phylogenetic analysis was performed with multiple alignments of protein-coding sequences, from the mitochondrial genomes, using PhyloSuite v1.2.2 (Zhang et al. Citation2020). A phylogenetic tree was constructed using Bayesian inference (BI) in MrBayes (Huelsenbeck and Ronquist Citation2001). The iTOL webtool (https://itol.embl.de) (Letunic and Bork Citation2007) was used to display the phylogenetic tree.
Results
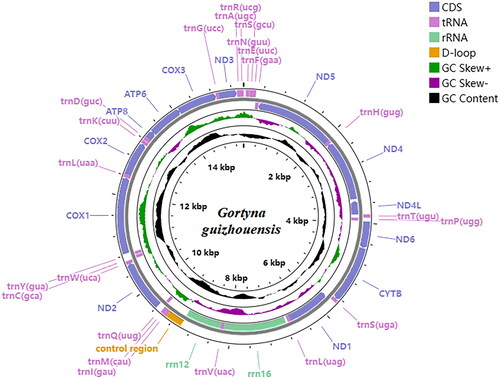
The coverage of the G. guizhouensis mitochondrial genome was 100%, and the reads in depth were evenly distributed across the mitochondrial genome (). The mitochondrial genome of G. guizhouensis was 15441 bp long (GenBank accession no. OM140690) and consisted of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and one control region (). The 16S RNA and 12S RNA genes were 1,359 bp and 785 bp long, respectively. The lengths of the 22 tRNA genes ranged from 64 bp (tRNA-Arg) to 72 bp (tRNA-Asp). The overall base composition of the mitogenome was estimated as follows: A 39.7%, T 39.5.4%, C 12.9% and G 8.0%, with a high A + T content of 79.1%. All PCGs started with the standard ATN codons apart from cox1, which started with CGA, and nd1, which started with TTG, Most of the PCGs terminated with the stop codon TAA, whereas cox2 and nd4 terminated with an incomplete stop codon T-.
Figure 2. Read coverage map across the G. guizhouensis mitochondrial genome. Circlize v0.4.16 was used for plotting.
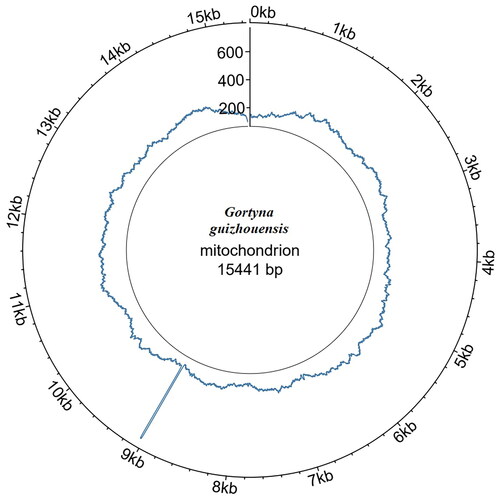
Figure 3. Location and arrangement of genes on the mitogenome of the G. guizhouensis. A circular mitochondrial genome map was drawn using the Proksee server (https://proksee.ca/).
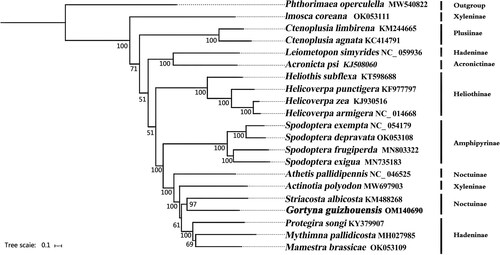
To gain insights into the volution of G. guizhouensis, we performed phylogenetic analysis using the complete mitogenomes of 13 PCGs from 21 Lepidoptera species. Of these species, 20 belonged to Noctuidae, and Phthorimaea operculella (Gelechiidae: Gelechiinae) was selected as the outgroup. Our analysis revealed that G. guizhouensis was closely related to Striacosta albicosta within the Noctuidae family. However, the monophyly of Hadeninae, Noctuinae, and Xyleninae could not be confirmed using the phylogenetic tree ().
Figure 4. Phylogenetic tree based on 13 mitochondrial protein-coding genes of 21 Lepidoptera species reconstructed by Bayesian inference (BI) method. Numbers on branches are Bootstrap support values (BS). The following sequences were used: MW540822 (Song et al. Citation2021), OK053111 (unpublished), KM244665 (unpublished), KC414791 (Gong et al. Citation2013), NC_059936 (unpublished), KJ508060 (unpublished), KT598688 (De Souza et al. Citation2016), KF977797 (Walsh Citation2016), KJ930516 (Perera et al. Citation2016), NC_014688 (Kômoto et al. Citation2011), NC_054179 (Li et al. Citation2021), OK053108 (unpublished), MN803322 (unpublished), MN735183 (unpublished), NC_046525 (Li et al. Citation2020), MW697903 (unpublished), KM488268 (Coates & Abel Citation2016), KY379907 (Zhao et al. Citation2022), MH027985 (unpublished), OK053109 (unpublished).
Discussion and conclusion
The mitochondrial genome of G. guizhouensis was 15,441 bp long and comprised 13 PCGs, 22 tRNA genes, two rRNA genes and one control region. In summary, the mitogenome of G. guizhouensis can provide the necessary molecular data for further phylogenetic and evolutionary analysis of Noctuidae. This study provides a mitochondrial genomic data for a new tobacco pest, that will be conducive to DNA classification and identification of pests in the future.
Ethical approval
Gortyna guizhouensis is a harmful insect that does not involve humans. This study does not need ethical approval or permissions to collect the sample.
Authors contributions
Meng Leng collected the specimen, analyzed the data and completed the drafting of the manuscript. Mao-fa Yang and Hai-yan Zhao were involved in the conception, design and revising it critically for intellectual content. Qiong Liu and Xiao-fei Yu collected and reared the larvae. All authors agreed the final approval to be published and agree to be accountable for all aspects of the work.
Acknowledgments
The authors sincerely thank Professor Hui-lin Han (Northeast Forestry University) for the identification of G. guizhouensis to species, and Hong-li He, Yan Jiang and Yan-fei Song (Guizhou University) for technical support. We would like to thank Editage (www.editage.cn) for English language editing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nih.gov under the accession no. OM140690. The associated BioProject, SRA and BioSample numbers are PRJNA826860, SRS12616393, and SAMN27585214, respectively.
Additional information
Funding
References
- Coates BS, Abel CA. 2016. The mitochondrial genome of the western bean cutworm, Striacosta albicosta (Lepidoptera: Noctuidae). Mitochondrial DNA B Resour. 1(1):487–488.
- De Souza BR, Tay WT, Czepak C, Elfekih S, Walsh TK. 2016. The complete mitochondrial DNA genome of a Chloridea (Heliothis) subflexa (Lepidoptera: Noctuidae) morpho-species. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4532–4533.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Gardiner T, Ringwood Z, Fairweather G, Perry R, Woodrow L. 2017. Introductions of two insect species threatened by sea-level rise in Essex, United Kingdom: fisher’s estuarine moth Gortyna borelii lunata (Lepidoptera: Noctuidae) and Mottled grasshopper Myrmeleotettix maculatus (Orthoptera: acrididae). Int Zoo Yearb. 51:69–78.
- Gong YJ, Wu QL, Wei SJ. 2013. Complete mitogenome of the Argyrogramma agnata (Lepidoptera: noctuidae). Mitochondrial DNA. 24(4):391–393.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Kômoto N, Yukuhiro K, Ueda K, Tomita S. 2011. Exploring the molecular phylogeny of phasmids with whole mitochondrial genome sequences. Mol Phylogenet Evol. 58(1):43–52.
- Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23:127–128.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Li Z, Wang W, Xiao Y, Zhang L. 2021. Characterization of the mitochondrial genome of Spodoptera exempta (Lepidoptera: Noctuidae) from South Africa. Mitochondrial DNA B Resour. 6(2):370–372.
- Li ZQ, Wu YP, Wang JP, Tw C, Wen JP. 2020. The complete mitochondrial genome of Athetis pallidipennis (Lepidoptera: Noctuidae). Mitochondrial DNA Part B. 5(2):1346–1348.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47:e63.
- Perera OP, Tk W, Luttrell RG. 2016. Complete mitochondrial genome of Helicoverpa zea (Lepidoptera: Noctuidae) and expression profiles of mitochondrial-encoded genes in early and late embryos. J Insect Sci. 16(1):40.
- Song YF, Xiao CG, Ye S, Zhang B, Yang MF, Liu JF. 2021. Complete mitogenome of Phthorimaea operculella (Lepidoptera: Gelechioidea: Gelechiidae). Mitochondrial DNA B Resour. 6(7):2088–2089.
- Walsh TK. 2016. Characterization of the complete mitochondrial genome of the Australian Heliothine moth, Australothis rubrescens (Lepidoptera: Noctuidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):167–168.
- Wu J, Yang M-F, Han H-L. 2022. A new species of the genus Gortyna Ochsenheimer, 1816 from China. Zootaxa. 5092(5):587–592. 5092.5.6
- Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhao JR, Zhang SP, Tang YY, Wang WZ, Tang BP, Liu QN, Yang RP. 2022. Comparative mitochondrial genome analysis of Mamestra configurata (Lepidoptera: noctuoidea: noctuidae) and other noctuid insects reveals conserved genome organization and phylogeny. Ann Entomol Soc Am. 115(3):304–313.
