Abstract
The beetle Triplax ainonia Lewis, 1877 is a serious pest of cultivated the mushroom Pleurotus ostreatus in China. The complete mitochondrial genome of this species was reported for the first time in this study. The mitogenome was 17,555 bp in length and had a base composition of 39.4% A, 36.1% T, 8.7% G and 15.3% C, which indicated that the base composition was AT-biased. Similar to other species of Coleoptera, the mitogenome of T. ainonia contained 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA unit genes, and a large noncoding region. Phylogenetic analysis based on mitogenomes suggested that the family Erotylidae was a monophyletic group.
Introduction
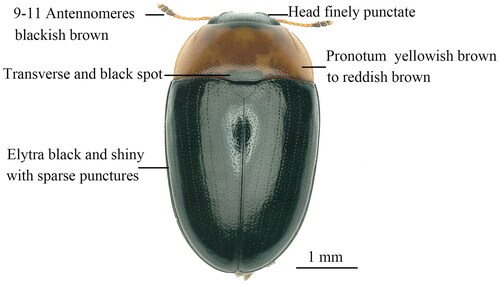
The genus Triplax Fabricius belongs to the tribe Tritomini of the subfamily Erotylinae, and the species of this genus are widely distributed in East Asia such as China, Korea and Japan (Jung and Park Citation2017; Jung Citation2019). Most species of the genus Triplax are fungivorous (Goodrich and Skelley Citation1993; Jung Citation2019). For example, we found that both the adults and the larvae of the species Triplax ainonia Lewis, 1877 can feed on the fruit bodies of the Pleurotus ostreatus and become a serious pest of this cultivated mushroom in China. This beetle species has the following typical morphological features: body elongate oval, mostly shiny and black; body length 2.82–4.86 mm, and width 1.63–2.67 mm; elytra bright black, head black; pronotum with two circular and black markings; antennae brown, but 9–11antennomeres dark brown; legs and abdomen orange. In this study, we report the complete mitochondrial genome of T. ainonia, representing the first mitogenome of the genus Triplax, and the results of this study will contribute to the species identification of this pest based on molecular data.
Materials and methods
The specimens of the Triplax ainonia were collected from Baijin Town, Huishui County, South Guizhou Autonomous Prefecture, Guizhou Province, China (26°6′32″N, 106°50′9″E) on 16 November 2021. The insect specimens used in this study were collected and identified by the author (Ben Hong). A specimen was deposited at the Institute of Entomology, Guizhou University (http://www.gzu.edu.cn/, Changqing Luo, [email protected]) under the voucher number GZU IE2021100101. The sample was preserved in 100% ethanol at −20 °C ().
Figure 1. Adult habitus of Triplax ainonia Lewis, 1877 (this photo was taken by Ben Hong, the first author of this article).
Genomic DNA of T. ainonia was extracted from one leg using the CTAB method (Reineke et al. Citation1998). DNA samples were stored at the Institute of Entomology, Guizhou University (http://www.gzu.edu.cn/, Changqing Luo, [email protected]) under the voucher number GZU E2021112801. A total amount of 1 µg DNA per sample was used as input material for the DNA library preparations. The DNA libraries were sequenced on Illumina NovaSeq 6000 platform (Illumina, USA), and 150 bp paired-end (PE) reads were generated. After quality assessment, a total of 38,710,978 raw reads were used to assemble the mitochondrial genome, and the assembly was annotated and visualized using MitoZ (Meng et al. Citation2019). Circularity was checked using the MITOS Web Server (Bernt et al. Citation2013, http://mitos.bioinf.uni-leipzig.de/), and manually revised based on the mitogenome of the species Neotriplax arisana which is closely related to T. ainonia. Thirteen protein-coding genes (PCGs) of the mitogenomes of five species, including the mitogenome of the species T. ainonia and all other known mitogenomes of the species belonging to the family Erotylidae of the Cucujoidea, and the mitogenomes of the species of four families (each family with two representative species) which also belong to the superfamily Cucujoidea were used for the phylogenetic analysis. One species of another superfamily (i.e. Sphaerioidea) of the Coleoptera was employed as the outgroup taxon. Except for the species T. ainonia, the complete mitochondrial sequences of the other 13 beetle species were downloaded from the NCBI (www.ncbi.nlm.nih.gov). Multiple sequence alignment was carried out using Clustal X 1.81 (Thompson et al. Citation1997). The maximum likelihood tree was reconstructed by MEGA X (Kumar et al. Citation2018) with 1000 bootstrap replications. The best model of evolution for the ML analyses was selected with MEGA X (Kumar et al. Citation2018), and the GTR + G+I was chosen as the final model of nucleotide substitutions.
Results
The complete mitochondrial genome of Triplax ainonia is a typical circular double-strand DNA molecule of 17,555 bp (GenBank accession no. OM084946). The mitogenome of this species contains 37 genes: 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), the small and large ribosomal RNA unit genes (rrnS and rrnL), and a large noncoding region (putative control region) (). The mitogenome of T. ainonia has a base composition of 39.4% A, 36.1% T, 8.7% G, 15.3% C.
Figure 2. Genome map of the mitogenome of T. ainonia. The outermost circle shows the arrangement of the genes: blue for the CDS, red for tRNAs, and orange for rRNAs.
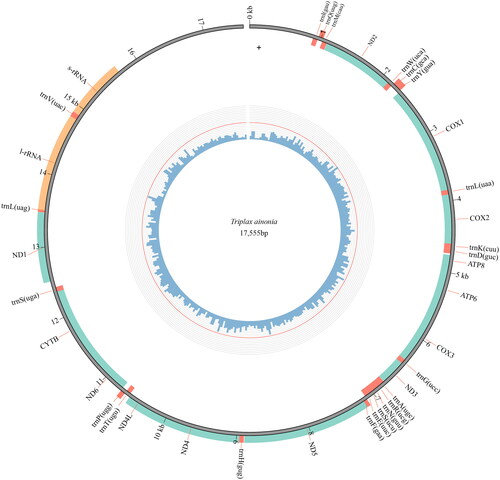
Twelve protein-coding genes use ATN (ATG, ATT, and ATA) as the start codons. Among those genes, six PCGs (cox2, cox3, nad4, nad6, cob, and atp6) start with ATG, five PCGs (cox1, atp8, nad3, nad5, and nad4l) start with ATT, and one PCG (nad2) starts with ATA. For the protein-coding gene nad1, TTG is used as a start codon. Eleven protein-coding genes stops with TAA, and the gene nad5 uses the TAG as a stop codon. However, the gene nad4 stop with an incomplete stop codon TA. The large noncoding region is 2,742 bp long, and located between rrnS and trnl. The 22 tRNAs are interspersed among ribosomal RNAs and protein-coding regions, and the size of these tRNAs varies from 64 bp (trnC, trnH, and trnT) to 70 bp (trnK and trnV). The rrnS is located between trnV and the noncoding region, and the rrnL is located between trnL and trnV. The rrnS and rrnL are 811 bp and 1329 bp long, respectively.
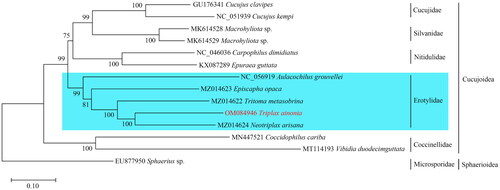
Phylogenetic relationships for 13 beetle species belonging to five families of the Cucujoidea were reconstructed (). The inferred phylogeny tree showed that the family Erotylidae which comprises the beetle species T. ainonia was a monophyletic group with 99% bootstrap probability (). Furthermore, the other four families formed a well-supported monophyletic clade, respectively (BP = 100%) ().
Figure 3. The phylogenetic tree resulting from maximum likelihood analysis. Numbers at the nodes represent bootstrap percentages. The following sequences were used: Cucujus clavipes GU176341 (Song et al. Citation2010); Cucujus kempi NC_051939, Macrophyliota sp. MK614528, Macrophyliota sp. MK614529 (Jin et al. Citation2020); Carpophilus dimidiatus NC_046036, Epuraea guttata KX087289 (Chen et al. Citation2020); Aulacochilus grouvellei NC_056919 (Liu et al. Citation2021); Episcapha opaca MZ014623, Tritoma metasobrina MZ014622, Neotriplax arisana MZ014624 (Liu et al. Citation2021); Coccidophilus cariba MN447521 (Nattier and Salazar Citation2019); Vibidia duoecimguttata MT114193 (Yan et al. Citation2020); Sphaerius sp. EU877950 (Sheffield et al. Citation2008).
Discussion and conclusions
This study determined the main features of the mitochondrial genome of the beetle Triplax ainonia. The gene composition of the mitogenome of this species resembles the mitogenomes of other beetle species belonging to Coleoptera (Liu et al. Citation2021; Xing et al. Citation2022). In addition, similar to the mtDNA sequences of other insects, the base composition of T. ainonia mitogenome is biased toward A + T bases (Yang et al. Citation2009). In the beetle T. ainonia, the gene nad4 stops with an incomplete stop codon, and this phenomenon of mitochondrial genes with immature stop codons have also been reported in other beetles (Liu et al. Citation2021; Xing et al. Citation2022).
The phylogenetic analysis showed that the Erotylidae formed a monophyletic clade, and the monophyly of this family was also confirmed by previous phylogenetic studies based on morphological and molecular characters (Lawrence and Newton Citation1982; Leschen et al. Citation2005; McElrath et al. Citation2015; Liu et al. Citation2021). In our results, the family Erotylidae was the sister to the other three families: Cucujidae + Silvanidae + Nitidulidae, and this topology was strongly supported. However, clarification of the phylogenetic relationships among families of the Cucujoidea need further phylogenetic studies which should be based on a much broader taxon sampling of this superfamily.
Ethical approval
The study was approved by the Guizhou University Subcommittee of Experimental Animal Ethics (approval no. EAE-GZU-2021-E015).
Authors’ contributions
Changqing Luo designed the study. Ben Hong and Yunxue Xiao performed the data analysis. Changqing Luo and Ben Hong drafted the manuscript. Yunxue Xiao critically reviewed the article. All authors approved the final version of the manuscript to be published and agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitochondrial genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OM084946. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA798621, SRR17731626, and SAMN25118147, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen X, Song Q, Huang M. 2020. Characterization of the complete mitochondrial genomes from two Nitidulid pests with phylogenetic implications. Insects. 11(11):779.
- Goodrich MA, Skelley PE. 1993. The pleasing fungus beetles of Illinois (Coleoptera: Erotylidae) Part II. Triplacinae. Triplax and Ischyrus. Trans Illinois State Acad Sci. 86(3, 4):153–171.
- Jin M, Zwick A, Ślipiński A, Marris JWM, Thomas MC, Pang H. 2020. A comprehensive phylogeny of flat bark beetles (Coleoptera: Cucujidae) with a revised classification and a new South American genus. Syst Entomol. 45(2):248–268.
- Jung BH, Park H. 2017. First record of two erotylid species of Triplax (Coleoptera: Erotylidae: tritomini) from Korea. J Species Res. 6(3):291–294.
- Jung BH. 2019. A new species and three new records of tribe Tritomini (Coleoptera: Erotylidae) in Korea. Int J Indust Entomol. 39(1):29–33.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lawrence JF, Newton AF. 1982. Evolution and classification of beetles. Annu Rev Ecol Syst. 13:261–290.
- Leschen RAB, Lawrence JF, Ślipiński SA. 2005. Classification of basal Cucujoidea (Coleoptera: polyphaga): cladistic analysis, keys and review of new families. Invertebr Syst. 19(1):17–73.
- Liu J, Lu W, Lv C, Shaw JJ, Li J. 2021. The complete mitochondrial genome of Aulacochilus grouvellei Achard, 1923 (Coleoptera: Erotylidae: Erotylinae) with a phylogenetic analysis of Cucujoidea. Mitochondrial DNA B Resour. 6(3):1150–1151.
- Liu J, Wang Y, Zhang R, Shi C, Lu W, Li J, Bai M. 2021. Three complete mitochondrial genomes of Erotylidae (Coleoptera: Cucujoidea) with higher phylogenetic analysis. Insects. 12(6):524.
- McElrath T, Robertson JA, Thomas MC, Osborne JT, Miller KB, McHugh J, Whiting MF. 2015. A molecular phylogenetic study of Cucujidae s.l. (Coleoptera: Cucujoidea). Syst Entomol. 40(4):705–718.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization.Nucleic Acids Res. 47(11):e63.
- Nattier R, Salazar K. 2019. Next-generation sequencing yields mitochondrial genome of Coccidophilus cariba Gordon (Coleoptera: Coccinellidae) from museum specimen. Mitochondrial DNA B Resour. 4(2):3780–3781.
- Reineke A, Karlovsky P, Zebitz CPW. 1998. Preparation and purification of DNA from insects for AFLP analysis. Insect Mol Biol. 7(1):95–99.
- Sheffield NC, Song H, Cameron SL, Whiting MF. 2008. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 25(11):2499–2509.
- Song H, Sheffield NC, Cameron SL, Miller KB, Whiting MF. 2010. When phylogenetic assumptions are violated: base compositional heterogeneity and among-site rate variation in beetle mitochondrial phylogenomics. Syst Entomol. l35(3):429–448.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882.
- Xing K, Chen K, Zhao XJ, Zhao F. 2022. The complete mitochondrial genome of Lixus subtilis Boheman, 1835 (Coleoptera, Curculionidae) and its phylogenetic implications. Mitochondrial DNA B Resour. 7(1):35–36.
- Yan J, Song P, Li Y, Tong X, Wang J, Liu D. 2020. Characterization of the complete mitochondrial genome of Vibidia duodecimguttata (Coleoptera: Coccinellidae). Mitochondrial DNA B Resour. 5(2):1565–1566.
- Yang L, Wei ZJ, Hong GY, Jiang ST, Wen LP. 2009. The complete nucleotide sequence of the mitochondrial genome of Phthonandria atrilineata (Lepidoptera: Geometridae). Mol Biol Rep. 36(6):1441–1449.
