Abstract
Lonicera caerulea var. edulis, known as “blue honeysuckle” or “Haskap,” is a deciduous shrub that belongs to the Caprifoliaceae family. Characterized by the high cold hardiness and high quality of fruit, it has become a novel cash crop in cold regions worldwide. The lack of available chloroplast (cp) genome information limits studies of its molecular breeding and phylogeny. Here, the complete cp genome of Lonicera caerulea var. edulis was assembled and characterized for the first time. It was 155,142 bp in length with 38.43% of GC content, including 23,841 bp inverted repeat regions (IRs), an 88,737 bp large single-copy region (LSC), and an 18,723 bp small single-copy region (SSC). A total of 132 genes, including 85 protein-coding genes, 8 rRNA genes, and 39 tRNA genes were annotated. Phylogenetic analysis indicated that L. caerulea var. edulis was closely related to L. tangutica. These data and results provide a valuable resource for the development of breeding tools and genetic diversity studies for L. caerulea.
Keywords:
Introduction
Lonicera caerulea var. edulis (Linn.) Turcz. ex Herd. 1864 (blue honeysuckle) is a deciduous shrub that produces blue-black edible berries with a high concentration of anthocyanins, phenolic acids, and polyphenols (Becker and Szakiel Citation2019). As a circumpolar species, its tree and flower are hardy to −50 °C and −7 °C, respectively (Plekhanova Citation2000, Hummer et al. Citation2012). Thus, its cultivation and breeding have undergone fast progress during the past two decades in the cold regions of the Northern Hemisphere, especially in Russia, China, and Canada (Huo et al. Citation2005, Gerbrandt et al. Citation2017).
Despite its importance, the phylogenetic position of L. caerulea in Lonicera is still less clear. It was designated as the only species of subsection Caeruleae in section Isika of the subgenus Chamaecerasus (Rehder Citation1903, Rehder Citation1913, Hara Citation1983, Wu and Hong Citation2011). Because of the lack of plant material or sequence data of L. caerulea, whether L. caerulea should be classified under Isika is still controversial, and the phylogenetic relationship between subsection Caeruleae and other subsections in section Isika remains unclear (Theis et al. Citation2008, Nakaji et al. Citation2015, Liu et al. Citation2018). Due to the genomic characteristics of nonrecombination, high conservation, and uniparental inheritance, the complete chloroplast (cp) genome has been accepted as a powerful tool for phylogenetic studies and molecular breeding (Bi et al. Citation2018). To date (August 2022), more than 40 versions of Lonicera cp assemblies have been released in GenBank, but L. caerulea has not been included.
Here, the cp genome of L. caerulea var. edulis was assembled and characterized for the first time and a plastome phylogeny of the genus Lonicera was reconstructed, providing useful genetic resources for the molecular breeding of L. caerulea and new insight into the phylogeny of Lonicera species.
Materials
Fresh leaves were obtained from a plant of L. caerulea var. edulis cultivated at the Horticulture Experimental Station (126.73 N, 45.74E) of Northeast Agricultural University (NEAU), Harbin, China. “LE02” was initially collected as a twig from the Lesser Khingan Mountains by Junwei Huo ([email protected]) in 2002 and was propagated by cutting. The botanical identification of “LE02” (Gui and Hu Citation2002, Zhang Citation2012) was accomplished by Professor Xiuju Wu ([email protected]). A specimen was deposited at NEAU (Junwei Huo, [email protected]) under the voucher number “LE-02”.
Methods
Total genomic DNA was extracted from 0.1 g of fresh leaves using a modified CTAB method (Doyle and Doyle Citation1986). A paired-end library with an average insert size of 350 bp was constructed using the MGIEasy PCR-Free DNA Library Prep Set and sequenced on the DNBSEQ™-T7 platform (MGI Tech Co., Ltd., Shenzhen, China). The raw reads were filtered using SOAPnuke (Chen et al. Citation2017) with the parameters “-lowQual = 20, -nRate = 0.005, qualRate = 0.5” (Supplementary File 1 and 2). The cp genome was assembled using GetOrganelle (Jin et al. Citation2020) with a k-mer set of “21, 45, 65, 85, 105” (Supplementary File 3 and Figure S1). The assembled genome was annotated using GeSeq (Tillich et al. Citation2017) and CPGView (Liu et al. Citation2022a). The 24 complete Lonicera cp genomes sequences were selected for phylogenetic analysis according to recent studies (). The 8 representative species in Caprifoliaceae and Adoxaceae were selected according to classical taxonomy (Wu and Hong Citation2011, The Angiosperm Phylogeny Group et al. 2016, Stevens Citation2017) and recent molecular studies (Theis et al. Citation2008, Nakaji et al. Citation2015, Bai et al. Citation2017, Liu et al. Citation2018, Wang et al. Citation2020, Wang et al. Citation2021). All of the sequences were aligned using MAFFT v7.505 (Katoh and Standley Citation2013) and then trimmed using trimAL (Capella-GUTIéRREZ et al. Citation2009) in the “-automated1” pattern. The maximum likelihood tree was constructed using IQ-TREE 2.2.0.3 with the best-fit model of “TVM + F + I + I+R6” and 1,000 ultrafast bootstraps (UFBoot) replicates for branch support (Minh et al. Citation2020).
Results and discussion
L. caerulea var. edulis () is one of the three diploid varieties (2n = 2x = 18) of L. caerulea and is a native species in northeastern Asia (Plekhanova Citation2000). Due to its widespread wild distribution and primitive karyotype, it is not only considered an ancestor of tetraploid L. caerulea but also has the potential to be an excellent material for ploidy breeding and a model system for gene function studies (Huo et al. Citation2005).
Figure 1. Tree (A) and fruit (B) morphologies of Lonicera caerulea var. edulis.
Note: location, Horticulture Experimental Station (126.73N, 45.74E), Northeast Agricultural University (NEAU), Harbin, China; botanical identification, Pro. Xiuju Wu; photograph, Chenqiao Zhu.

The complete cp genome of L. caerulea var. edulis is 155,142 bp in length, with 38.43% of GC content. It includes a pair of IRs of 23,841 bp, which separate the LSC of 88,737 bp from the SSC of 18,723 bp (). A total of 132 genes were annotated, including 85 protein-coding genes (PCGs), 8 rRNA genes (rRNAs), and 39 tRNA genes (tRNAs). Among them, 5 PCGs and 8 tRNAs were repeated in IRs, ten PCGs and 7 tRNAs contained one intron, pafI contained two introns, and rps12 underwent trans-splicing (, Figure S2 and ).
Figure 2. Feature map of Lonicera caerulea var. edulis chloroplast genome.
From the center outward, the first track shows the dispersed repeats, which consist of direct (red arc) and Palindromic (green arc). The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors (Black: complex repeat; Green: 1-unit repeat). The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content is plotted on the fifth track. The annotated genes are shown on the sixth track (codon usage bias is displayed in the parenthesis) and their functional classifications are color-coded in the bottom left corner. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively.
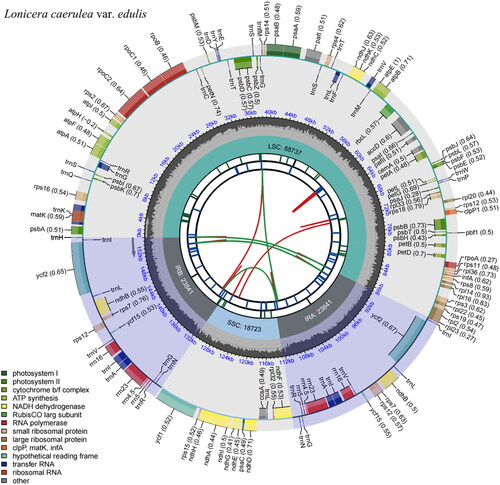
Figure 3. Phylogenetic position of Lonicera caerulea var. edulis inferred from maximum likelihood (ML) based on 33 complete chloroplast genomes.
The subclassification of the Lonicera species examined in this study is according to Rehder (Citation1903, Citation1913), and the representative colors of the subsections are shown on the left diagram. The subfamily classification was labeled according to Wang et al. (Citation2020). The Numbers shown next to the nodes are bootstrap support values based on 1,000 replicates. The following sequences were used: MW795591 (Liu et al. Citation2022b), MZ779026 (Zhang et al. Citation2022), MZ241298 (Jiang et al. Citation2021), NC060471 (Wei et al. Citation2021), MW186760 (Yu et al. Citation2021), MW186761 (Gu et al. Citation2021), MH579750 (Hu et al. Citation2018), MZ901373 (Yang et al. Citation2022), OK393707 (Chen et al. Citation2022), MW296954 (Gu et al. Citation2022), MW340876 (Yuan et al. Citation2021), MN256451 (Jia et al. Citation2020), MW242826, MK176510 (Liu et al. Citation2018), OL457164 (Mo et al. Citation2022), MH681655 (Zhu et al. Citation2019), MK176512 (Liu et al. Citation2018), MH028743 (Kang et al. Citation2018), OP345475 (assembled in the present study), MZ962399 (Wang et al. Citation2022), MK176511 (Liu et al. Citation2018), MG738668 (Fan et al. Citation2018), MG738669 (Fan et al. Citation2018), MH028740 (Kang et al. Citation2018), MG738667 (Fan et al. Citation2018), NC045051 (Wang et al. Citation2020), NC042190, NC029874 (Bai et al. Citation2017), NC045063 (Wang et al. Citation2020), NC045046 (Wang et al. Citation2020), MN524626 (Wang et al. Citation2020), NC033878 (Fan et al. Citation2018), NC032296 (Huang and Cronk Citation2015).
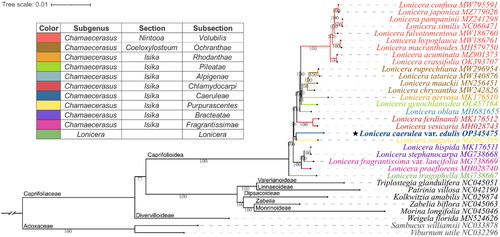
Table 1. Gene annotation summary of Lonicera caerulea var. edulis chloroplast genome.
Phylogenetic analysis () showed that the outgroup (Adoxaceae) was located at the base of the tree, and Caprifoliaceae fell into 7 clades with a highly supported topology of (Weigela, (Lonicera, ((Triplostegia, Patrinia), (Kolkwitzia, (Zabelia, Morina))))), which highly supports the molecular taxonomy of Caprifoliaceae (Wang et al. Citation2020). The Lonicera clade is divided into two main clades, namely subgenus Lonicera (L. tragophylla) and subgenus Chamaecerasus (other 24 Lonicera species). However, no hierarchical topology was found among or inside sections Coeloxylosteum and Isika. L. caerulea var. edulis (subsection Caerueae) clustered with L. tangutica (subsection Purpurascentes) in the same branch on the clade of section Isika, indicating their close phylogenetic relationship. These results challenge the subclassification of the subgenus Chamaecerasus (Rehder Citation1903, Rehder Citation1913), suggesting that multi-data molecular phylogenetic studies are needed to resolve species relationships and guide taxonomic revisions for the genus Lonicera.
Conclusion
The cp genome characterization and phylogenetic analysis of L. caerulea provide useful genetic resources and new insights for future molecular breeding and phylogenetic studies.
Ethical statement
Appropriate permissions were granted by NEAU before sampling. The initial wild collection followed the “Regulations of the People”s Republic of China on the Protection of Wild Plants guidelines” (http://www.forestry.gov.cn) and complied with permission from the Heilongjiang Forestry and Grassland Bureau (http://lyhcyj.hlj.gov.cn/).
Authors” contributions
JWH. and CQZ. designed this research. CQZ., LJZ., XYS., QF., YZ., SLL., YL., MY., and DQ. partially participated in the experiment and data analysis. CQZ. drafted the manuscript.
Supplemental Material
Download MS Word (14.8 MB)Disclosure statement
No potential conflict of interest was reported by all the coauthors.
Data availability statement
The cp assembly in this study is openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under accession no. OP345475. The associated BioProject, SRA, and BioSample numbers are PRJNA870700, SRR21133392, and SAMN30383140 respectively.
Additional information
Funding
References
- Bai G-Q, Zhou T, Zhao J-X, Li W-M, Han G-J, Li S-F. 2017. The complete chloroplast genome of Kolkwitzia amabilis (Caprifoliaceae), an endangered horticultural plant in China. Mitochondrial DNA Part A. 28(2):296–297.
- Becker R, Szakiel A. 2019. Phytochemical characteristics and potential therapeutic properties of blue honeysuckle Lonicera caerulea L. (Caprifoliaceae). J Herb Med. 16:100237.
- Bi Y, Zhang M-F, Xue J, Dong R, Du Y-P, Zhang X-H. 2018. Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Sci Rep. 8(1):1184.
- Capella-GUTIéRREZ S, Silla-MARTíNEZ JM, GABALDóN T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, et al. 2017. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 7(1):1–6.
- Chen C, Qu D-H, Shan F-Q, Jin Z-X, Sun Z-S. 2022. Complete chloroplast genome of Lonicera crassifolia Batalin (Caprifoliaceae) and its phylogenetic implications. Mitochondrial DNA Part B. 7(5):732–734.
- Doyle J, Doyle J. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19(1):11–15.
- Fan W-B, Wu Y, Yang J, Shahzad K, Li Z-H. 2018. Comparative chloroplast genomics of dipsacales species: insights into sequence variation, adaptive evolution, and phylogenetic relationships. Front Plant Sci. 9:689.
- Gerbrandt E, Bors B, Chibbar R, Baumann T. 2017. Spring phenological adaptation of blue honeysuckle (Lonicera caerulea L.) foundation germplasm in a temperate climate. Can J Plant Sci. 98(3):569–581.
- Gu L, Hou Y, Wang G, Liu Q, Ding W, Weng Q. 2022. Characterization of the chloroplast genome of Lonicera ruprechtiana Regel and comparison with other selected species of Caprifoliaceae. PLOS One. 17(1):e0262813.
- Gui M, Hu Y. 2002. Cultivation biology of small berry (in Chinese). Beijing, China: China Agriculture Press.
- Gu L, Wu Q, Yi Y, Yu Z. 2021. The complete chloroplast genome of Lonicera hypoglauca Miq (Caprifoliaceae: Dipsacales) from Guangxi. Mitochondrial DNA Part B. 6(2):450–451.
- Hara H. 1983. A revision of Caprifoliaceae of Japan with reference to allied plants in other districts and the Adoxaceae. Ginkgoana. 5:331–355.
- Hu H, Liu J, An J, Wang M, Wang Q. 2018. Characterization of the complete chloroplast genome of Lonicera macranthoides. Mitochondrial DNA Part B. 3(2):1000–1001.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Hummer K, Pomper K, Postman J, Graham C, Stover E, Mercure E, Aradhya M, Crisosto C, Ferguson L, Thompson M, et al. 2012. Emerging Fruit Crops. In: Badenes, M. and Byrne, D. eds. Fruit Breeding. Boston, MA.: Springer, p. 97–147.
- Huo J, Sui W, Yang G, Yu Z. 2005. Review of study on germplasm resources of blue honeysuckle (Lonicera caerulea L.). Acta Horticulturae Sinica. 32(1):159–164.
- Jia G, Wang H, Yu P, Li P. 2020. The complete chloroplast genome of the Lonicera maackii (Caprifoliaceae), an ornamental plant. Mitochondrial DNA Part B. 5(1):560–561.
- Jiang C, Wu S, Feng X, Yang C, Yu Z. 2021. The complete chloroplast genome of Lonicera pampaninii Levl. and its phylogenetic analysis. Mitochondrial DNA Part B. 6(10):3025–3027.
- Jin J-J, Yu W-B, Yang J-B, Song Y, Depamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kang K-B, Kang S-J, Kim M-S, Lee D-Y, Han S-I, Kim T-B, Park J-Y, Kim J, Yang T-J, Sung S-H. 2018. Chemical and genomic diversity of six Lonicera species occurring in Korea. Phytochemistry. 155:126–135.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu M-L, Fan W-B, Wang N, Dong P-B, Zhang T-T, Yue M, Li Z-H. 2018. Evolutionary analysis of plastid genomes of seven Lonicera L. species: implications for sequence divergence and phylogenetic relationships. IJMS. 19(12):4039.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2022a. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 0:1–11.
- Liu S, Zhou L, Huang J, Zeng H, Qiao Z, Li Y, Zhang G. 2022b. Comparative analysis of the complete chloroplast genome sequences of four origin plants of Lonicerae Flos (Lonicera; Caprifoliaceae). Phyton-Int J Exp Bot. 91:1503–1516.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, VON Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Mo R-J, Wang H-X, Chen D-J, Zhu Z-X, Qiao W, Wang H-F. 2022. Complete plastome sequence of Lonicera gynochlamydea Hemsl. (Caprifoliaceae). Mitochondrial DNA Part B. 7(7):1380–1381.
- Nakaji M, Tanaka N, Sugawara T. 2015. A molecular phylogenetic study of Lonicera L.(Caprifoliaceae) in Japan based on chloroplast DNA sequences. Acta Phytotax Geobot. 66(3):137–151.
- Plekhanova M. 2000. Blue honeysuckle (Lonicera caerulea L.)-a new comercial berry crop for temperate climate: genetic resources and breeding. Acta Hortic. (538 (Vol):159–164.
- Rehder A. 1903. Synopsis of the genus Lonicera. Ann. Rep. Missouri Bot. Gard. 14:27–232.
- Rehder A. 1913. Caprifoliaceae. Sargent C S. Plantae Wilsonianae. Cambridge: Cambridge University Press, 1, p. 128.
- Stevens PF. 2017. Angiosperm phylogeny website, version 14. [online]. University of Missouri–St Louis and Missouri Botanical Garden. St. Louis, USA. Available from: http://www.mobot.org/MOBOT/research/APweb/. [Accessed 4 July 2017].
- The Angiosperm Phylogeny Group, Chase MW, Christenhusz MJM, Fay MF, Byng JW, Judd WS, Soltis DE, Mabberley DJ, Sennikov AN, Soltis PS, Stevens PF, 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181(1):1–20.
- Theis N, Donoghue MJ, Li J. 2008. Phylogenetics of the Caprifolieae and Lonicera (Dipsacales) based on nuclear and chloroplast DNA sequences. Issn: 0363–6445. 33(4):776–783.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang X, He J, Su X, Xu B, Liu Y, Tang Y, Sun S, Li P, Zhao C. 2022. Characterization of the complete chloroplast genome of Lonicera tangutica (Caprifoliaceae), an ornamental and medicinal plant in China. Mitochondrial DNA Part B. 7(3):535–536.
- Wang H-X, Liu H, Moore MJ, Landrein S, Liu B, Zhu Z-X, Wang H-F. 2020. Plastid phylogenomic insights into the evolution of the Caprifoliaceae s.l. (Dipsacales). Mol Phylogenet Evol. 142:106641.
- Wang H-X, Morales-Briones DF, Moore MJ, Wen J, Wang H-F. 2021. A phylogenomic perspective on gene tree conflict and character evolution in Caprifoliaceae using target enrichment data, with Zabelioideae recognized as a new subfamily. J Syst Evol. 59(5):897–914.
- Wei X-M, Hu Y, Wei K-H, Wu Q-H, Huang Y, Wei F. 2021. Characterization of the complete chloroplast genome of Lonicera similis (Caprifoliaceae). Mitochondrial DNA Part B. 6(10):3067–3069.
- Wu Z, Hong D. 2011. Flora of China (English version). Beijing, China: Science Press.
- Yang C, Jiang C, Wu S, Feng X, Yu Z. 2022. The complete chloroplast genome of Lonicera acuminata Wall. and its phylogenetic analysis. Mitochondrial DNA Part B. 7(5):807–809.
- Yu Z, Yi Y, Gu L. 2021. The complete chloroplast genome of Lonicera fulvotomentosa Hsu et S. C. Cheng and its phylogenetic analysis. Mitochondrial DNA Part B. 6(3):842–843.
- Yuan W, Ma Y, He S, Chang D, He Y. 2021. Characterization of the complete chloroplast genome of Lonicera tatarica L. (Caprifoliaceae). Mitochondrial DNA Part B. 6(7):1871–1872.
- Zhang J, Liu H, Xu W, Wan X, Zhu K. 2022. Complete chloroplast genome sequences of Lonicera japonica cv. Damaohua: characterization and comparative and phylogenetic analyses. PREPRINT (Version 1) available at Research Square.
- Zhang L. 2012. [Study on the morphological diversity and fruit quality of blue honeysuckle (Lonicera caerulea L]. ) (in Chinese)[Master”s thesis. Northeast Agricultural University].
- Zhu Y-X, Wu Y-M, Shen X-L, Tong L, Xia X-F, Mu X-Y, Zhang Z-X. 2019. The complete chloroplast genome of Lonicera oblata, a critically endangered species endemic to North China. Mitochondrial DNA Part B. 4(2):2337–2338.
