Abstract
Momordica cochinchinensis (Lour.) Spreng. is an important medicinal plant that is used to treat various diseases in South and Southeast Asia. In this study, the complete plastome of M. cochinchinensis was sequenced and found to exhibit a total length of 158,955 bp, with a large single copy (LSC) region of 87,924 bp and a small single copy (SSC) region of 18,479 bp, as well as with two inverted repeats (IRs) that were both 26,726 bp in length. In total, 129 genes were detected, comprising 86 protein-encoding genes, 8 ribosomal RNA (rRNA) genes, and 35 transfer RNA (tRNA) genes. Furthermore, the inferred phylogenetic tree confirmed that M. cochinchinensis belongs to the genus Momordica in the Cucurbitaceae family. The research results will be used for authenticating M. cochinchinensis plant materials and for analyzing the genetic diversity and phylogenetic relationships in Momordica.
Introduction
Momordica cochinchinensis (Lour.) Spreng. 1826, commonly known as Gac fruit, sweet gourd, baby jackfruit, or cochinchin gourd (Bootprom et al. Citation2015), is a perennial dioecious cucurbit plant that originated in South and Southeast Asia and that is widely sold for dietary and medicinal purposes (Vuong et al. Citation2006). Momordica cochinchinensis is highly rich in lycopene and beta-carotene, vitamin E, fatty acids, flavonoids, phenolic acids, and trypsin inhibitors (Chuyen et al. Citation2015). These phytochemicals are associated with many significant pharmacological activities, such as provitamin A, antioxidant, antimicrobial, antiulcer, and anticancer activities (Jayanthi et al. Citation2020). The pulp of the seed or aril of the ripe fruit is usually utilized as a natural colorant and food additive because of its bright red color and rich nutritional content (Bootprom et al. Citation2013). The genetic diversity of Gac fruit is also important for germplasm exploration and selective breeding; unfortunately, there is a lack of genetic information about the underutilized crop (Bootprom et al. Citation2015; Jayanthi et al. Citation2020). The complete plastome of many plants has been sequenced, and DNA molecular markers have been developed and used for the identification of species and phylogenetic analysis (Li et al. Citation2022). Until now, there has been no research on the use of the whole plastome of M. cochinchinensis as a molecular genetic resource. Therefore, the current research was conducted to publish the whole chloroplast genome of M. cochinchinensis in order to enhance the molecular investigation of germplasm, genetic diversity, and phylogenetic relationships.
Materials and methods
Momordica cochinchinensis seeds were collected at Fangchenggang (21°46′8.87″N, 108 21′ 12.31″), Guangxi, China, and seedlings of M. cochinchinensis (voucher number: QZ06MC, ) were grown at the ecological garden of Jiangxi Agricultural University, Nanchang, China (contact person: Qianglong Zhu, [email protected]). Young and healthy plant leaves were sampled, and total genomic DNA (gDNA) was isolated according to the modified cetyltrimethylammonium bromide (CTAB) method (Porebski et al. Citation1997). Approximately, 15 μg of gDNA was isolated and delivered to the Beijing Genomics Institute (BGI) for genomic sequencing by using the BGISEQ-500 Platform (Shenzhen, China). Nearly 0.5 gigabytes (Gb) of clean paired reads were obtained after checking the sequence data quality. Plasmidspades.py was used to assemble the draft genome sequence (Bankevich et al. Citation2012), and the top scaffolds with high coverage and length for the plastome were then extracted, ordered, and combined into a plastome sequence draft according to the reference plastome of Momordica charantia (NC_036807.1). GapCloser was used to repair the gaps within the plastome sequence draft, and the integrality and quality of the plastome sequence were checked and improved by reference-guided mapping using Burrows–Wheeler Aligner (BWA), SAMtools and Integrative Genomics Viewer (IGV). The average and minimum read mapping depths of the assembled genome were 58× and 18× (Figure S1), respectively. Finally, the genes in the plastome sequence were annotated using GeSeq and CPGAVAS2 (Chang et al. Citation2012; Tillich et al. Citation2017). The annotated results were manually reviewed and corrected with Sequin software. The circular M. cochinchinensis plastome map was drawn using CPGView (Liu et al. Citation2023). For phylogenetic analysis, complete plastome sequences were aligned by using Multiple Alignment using Fast Fourier Transform (MAFFT, v7.463) (Rozewicki et al. Citation2019), and a phylogenetic tree was constructed based on the sequence alignment by Molecular Evolutionary Genetics Analysis (MEGA) (v11) with the maximum-likelihood (ML) method (Tamura et al. Citation2021).
Results
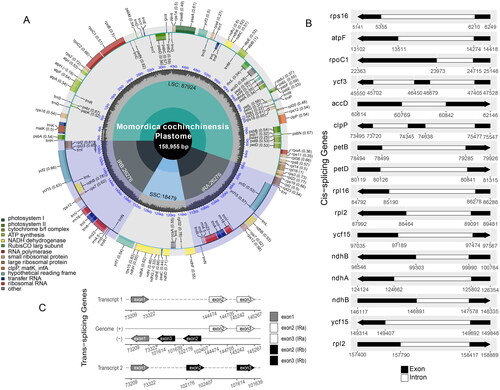
The length of the complete M. cochinchinensis plastome (ON597626) was 158,955 bp with 36.22% GC content (), contained the LSC (87,924 bp) and SSC (18,479 bp) regions and two IRs that were both 26,726 bp in length, and showed a typical quadripartite structure. However, a total of 129 genes were identified as being distributed in different regions of the plastome, including 86 protein-encoding genes, 35 tRNA genes, and 8 rRNA genes. Of these genes, 13 protein-encoding and 7 tRNA genes harbored at least two exons.
Figure 2. The complete plastome map of M. cochinchinensis. (A) The circle map of complete plastome of M. cochinchinensis. Different colored boxes on the outer circle represent genes. The clockwise and counter-clockwise genes transcribed are drawn inside and outside of the circle, respectively. The gray color region inside the inner circle indicates the GC content, and the quadripartite structure (LSC, SSC, IRa, and IRb) is drawn on the inner circle, respectively. (B) The black-white arrow showed the cis-spliced genes, and (C) the black-grey-white arrow showed trans-spliced genes (rps12).
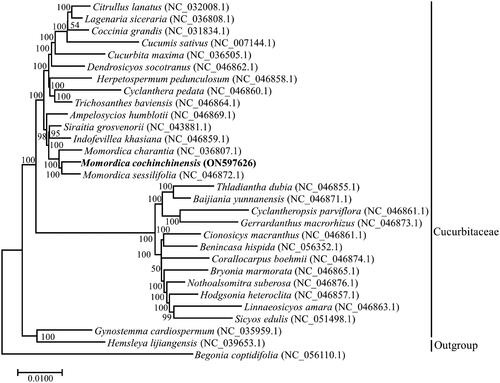
To date, there are two complete plastomes (Momordica charantia and Momordica sessilifolia) in the genus Momordica (Bellot et al. Citation2020), and approximately, 62 complete plastomes of Cucurbitaceae have been deposited in the NCBI Genome database; these complete plastomes represent 27 genera. To confirm the phylogenetic status of M. cochinchinensis, 30 complete plastome sequences representing the different genera of Cucurbitaceae and an outgroup species (Begonia coptidifolia) were used to infer a phylogenetic tree. The inferred phylogenetic tree suggested that M. cochinchinensis is closely related to M. charantia and M. sessilifolia, and that it belonged to Momordica in the Cucurbitaceae family ().
Figure 3. Phylogenetic tree indicated the relationship of M. cochinchinensis with other 29 species within the Cucurbitaceae, Begonia coptidifolia was regarded as the Outgroup. The complete plastomes were applied to infer the phylogenetic tree with Maximum-likelihood method and 1000 as bootstrap value, number above branches represent bootstrap value. The following sequences with GenBank accession were used: Citrullus lanatus NC_032008.1 (Zhu et al. Citation2016), Lagenaria siceraria NC_036808.1, Coccinia grandis NC_031834.1 (Sousa et al. Citation2016), Cucumis sativus NC_007144.1 (Plader et al. Citation2007), Cucurbita maxima NC_036505.1, Dendrosicyos socotranus NC_046862.1 (Bellot et al. Citation2020), Herpetospermum pedunculosum NC_046858.1 (Bellot et al. Citation2020), Cyclanthera pedata NC_046860.1 (Bellot et al. Citation2020), Trichosanthes baviensis NC_046864.1 (Bellot et al. Citation2020), Ampelosycios humblotii NC_046869.1 (Bellot et al. Citation2020), Siraitia grosvenorii NC_043881, Indofevillea khasiana NC_046859.1 (Bellot et al. Citation2020), Momordica charantia NC_036807.1, Momordica sessilifolia NC_046872.1 (Bellot et al. Citation2020), Thladiantha dubia NC_046855.1 (Bellot et al. Citation2020), Baijiania yunnanensis NC_046871.1 (Bellot et al. Citation2020), Cyclantheropsis parviflora NC_046861.1 (Bellot et al. Citation2020), Gerrardanthus macrorhizus NC_046873.1 (Bellot et al. Citation2020), Cionosicys macranthus NC_046861.1 (Bellot et al. Citation2020), Benincasa hispida NC_056352.1, Corallocarpus boehmii NC_046874.1 (Bellot et al. Citation2020), Bryonia marmorata NC_046865.1 (Bellot et al. Citation2020), Nothoalsomitra suberosa NC_046876.1 (Bellot et al. Citation2020), Hodgsonia heteroclita NC_046857.1 (Bellot et al. Citation2020), Linnaeosicyos amara NC_046863.1 (Bellot et al. Citation2020), Sicyos edulis NC_051498.1 (Cui et al. Citation2021), Gynostemma cardiospermum NC_035959.1, Hemsleya lijiangensis NC_039653.1 (Zhang et al. Citation2018), Begonia coptidifolia NC_056110.1 (Wang et al. Citation2021).
Discussion and conclusion
The complete plastome of M. cochinchinensis was first sequenced and found to exhibit a total length of 158,955 bp. A total of 129 genes were annotated, comprising 86 protein-encoding genes, 8 rRNA genes, and 35 tRNA genes. The genome size and gene content are not significantly different from those of most chloroplast genomes or plastomes in Cucurbitaceae (Zhang et al. Citation2018; Bellot et al. Citation2020). Furthermore, the inferred phylogenetic tree analysis confirmed that M. cochinchinensis belonged to the genus Momordica in the Cucurbitaceae family, which further supported earlier reported studies related to the phylogenetic relationship of M. cochinchinensis (Chomicki et al. Citation2020; Ghosh et al. Citation2021). The other species of different genera have been distinctly separated, but the topology of the inferred phylogeny is different from those of recently published cucurbit phylogenies that are based on filtered plastid and nuclear loci (Bellot et al. Citation2020), suggesting that further research on phylogenetic relationships in Momordica is necessary and should entail combining molecular markers from plastid and nuclear genomes. Our research results could be used for authenticating M. cochinchinensis and analyzing the genetic diversity and phylogenetic relationships in Momordica.
Ethical approval
No permission was necessary in this study for the sample collection. Momordica cochinchinensis (Lour.) Spreng. is widely grown in the Yangtze River basin and areas of the South, and is not on the List of National Key Protected Wild Plants.
Author contributions
L. J. Cai and R. Pan analyzed the sequence data and drafted the paper; Q. Zeng, X. Y. Zhang and R. B. Zeng collected the specimen material and conducted the experiment; Q. L. Zhu contributed to the conception and designing of this work; All authors carefully read, revised, and approved the final manuscript to be published. Thanks for Dr. Sikandar Amanullah performed reviewing, and editing of the paper.
Supplemental Material
Download MS Word (30.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The supported data for the findings of this study are publicly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/ON597626.1/, reference number ON597626. The clean reads used in this study have been deposited in the CNGBdb database (https://db.cngb.org/). The associated project, sample, and experiment numbers are CNP0003312, CNS0582170, and CNX0484017, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bellot S, Mitchell TC, Schaefer H. 2020. Phylogenetic informativeness analyses to clarify past diversification processes in Cucurbitaceae. Sci Rep. 10(1):488.
- Bootprom N, Songsri P, Suriharn K, Chareonsap P, Sanitchon J, Lertrat K. 2013. Molecular diversity among selected Momordica cochinchinensis (Lour.) spreng accessions using rapid markers. Sabrao J Breed Genet. 44(2):406–417.
- Bootprom N, Songsri P, Suriharn K, Lomthaisong K, Lertrat K. 2015. Genetics diversity based on agricultural traits and phytochemical contents in spiny bitter gourd: (Momordica cochinchinensis (lour.) spreng). Sabrao J Breed Genet. 47(3):278–290.
- Chang L, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Chomicki G, Schaefer H, Renner SS. 2020. Origin and domestication of Cucurbitaceae crops: insights from phylogenies, genomics and archaeology. New Phytol. 226(5):1240–1255.
- Chuyen HV, Nguyen MH, Roach PD, Golding JB, Parks SE. 2015. Gac fruit (Momordica cochinchinensis Spreng.): a rich source of bioactive compounds and its potential health benefits. Int J Food Sci Technol. 50(3):567–577.
- Cui H, Zhu Z, Lu Z, Ding Z, Zhang C, Luan F. 2021. The complete chloroplast genome sequence of the Sechium edule. Mitochondrial DNA B Resour. 6(1):97–98.
- Ghosh I, Saha PS, Bhowmick BK, Jha S. 2021. A phylogenetic analysis of Momordica (Cucurbitaceae) in India based on karyo-morphology, nuclear DNA content and rDNA ITS1-5.8S-ITS2 sequences. Protoplasma. 258(2):347–360.]
- Jayanthi P, Chew H, S S, Chew BL, Ong MT. 2020. Momordica cochinchinensis Spreng (Gac fruit): an abundant source of nutrient, phytochemicals and its pharmacological activities. Aust J Crop Sci. 14:1844–1854.
- Li B, Liu T, Ali A, Xiao Y, Shan N, Sun J, Huang Y, Zhou Q, Zhu Q. 2022. Complete chloroplast genome sequences of three Aroideae species (Araceae): lights into selective pressure, marker development and phylogenetic relationships. BMC Genomics. 23(1):218–234.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 0:1–11.
- Plader W, Yukawa Y, Sugiura M, Malepszy S. 2007. The complete structure of the cucumber (Cucumis sativus L.) chloroplast genome: its composition and comparative analysis. Cell Mol Biol Lett. 12(4):584–594.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter. 15(1):8–15.
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10.
- Sousa A, Bellot S, Fuchs J, Houben A, Renner SS. 2016. Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y chromosome reveals distinct Y centromeres. Plant J. 88(3):387–396.
- Tamura K, Stecher G, Kumar S. 2021. MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):w6–w11.
- Vuong LT, Franke AA, Custer LJ, Murphy SP. 2006. Momordica cochinchinensis Spreng. (Gac) fruit carotenoids reevaluated. J Food Compos Anal. 19(6):664–668.
- Wang Z, Liu TH, Cao HL. 2021. The complete chloroplast genome sequence of Begonia coptidifolia. Mitochondrial DNA B Resour. 6(2):548–549.
- Zhang X, Zhou T, Yang J, Sun J, Ju M, Zhao Y, Zhao G. 2018. Comparative analyses of chloroplast genomes of Cucurbitaceae species: lights into selective pressures and phylogenetic relationships. Molecules. 23:2165.
- Zhu Q, Cui H, Zhao Y, Gao P, Liu S, Wang P, Luan F. 2016. The complete chloroplast genome sequence of the Citrullus lanatus L. Subsp. Vulgaris (Cucurbitaceae). Mitochondrial DNA B Resour. 1(1):943–944.

