Abstract
Isoetes orientalis is an endangered hexaploidy species of Isoetaceae in China and the complete chloroplast genome of this species has not been reported. In the present study, a complete chloroplast genome of Isoetes orientalis (Isoetaceae) was assembled and annotated. This chloroplast genome has a circular structure of 145,504 bp in length, comprising a pair of inverted repeat (IR) regions of 13,207 bp each, a large single-copy (LSC) region of 91,864 bp, and a small single-copy (SSC) region of 27,226 bp. The chloroplast genome contains 136 genes, including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis showed that I. orientalis was closely related to I. sinensis. These results provide additional resources for future studies on Isoetes from China and across the globe.
Introduction
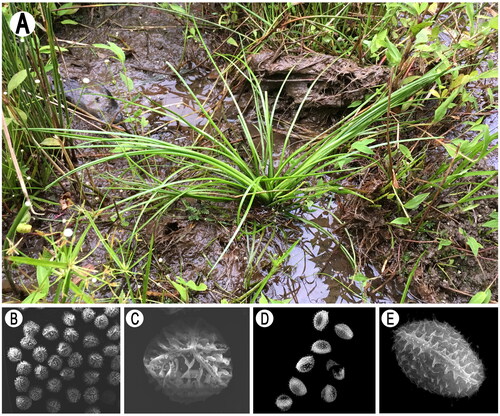
Isoetes orientalis Hong Liu & Q. F. Wang, an endangered hexaploid quillwort belonging to Isoetaceae, was reported in 2005 (Liu et al. Citation2005) and previously identified as I. sinensis (Pang et al. Citation2003; Ye and Li Citation2003; Chen et al. Citation2004; Kang et al. Citation2005), because this species has 66 chromosomes and spore features are different from I. sinensis, it was identified as new species. Isoetes orientalis is an emergent aquatic plant growing in the marshes of Songyang County of Zhejiang Province, China (Liu et al. Citation2005). Megaspores of this species have cristate-reticulate ornamentation, while microspores have echinate-tuberculate ornamentation (Liu et al. Citation2005, Citation2008) ().
Using NGS to infer evolutionary relationships with genomic-scale datasets allows researchers to resolve the branching order within rapid radiations and obtain a more robust phylogenetic framework (Wei et al. Citation2017; Wei and Zhang Citation2020). Plastid genome (plastome) DNA sequences have been extensively used in recent plant molecular systematics because of their mode of uniparental inheritance and high content of informative loci (Givnish et al. Citation2010; Jansen et al. Citation2011; Ruhfel et al. Citation2014; Givnish et al. Citation2015; Lu et al. Citation2015; Ross et al. Citation2016; Schafran et al. Citation2018). As an endangered species, Isoetes orientalis is also the only hexaploidy species in China, which would be very important in the speciation path of Isoetaceae of China even East Asia. Therefore, it will be significant to know its complete chloroplast genome sequence.
Materials and methods
In the present study, fresh leaf material was obtained from the collection sites of Andaihou Village, Songyang County, Lishui City, China (119.273422 E, 28.271808 N) () and dried with silica. Specimens (voucher no.: Yufeng Gu Fern08748) were deposited at the Herbarium of the National Orchid Conservation Center (NOCC). Silica-dried material was sent to Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China) for DNA extraction and sequencing, performed on an Illumina HiSeq X Ten platform (Illumina, San Diego, CA). The plastid genome was assembled using GetOrganelle v1.7.5 (Jin et al. Citation2018) using default parameters, and the results viewed and edited by Bandage v0.8.1 (Wick et al. Citation2015). The assembled chloroplast genome was annotated by Geneious Prime 2021.0.3 (https://www.geneious.com) (Kearse et al. Citation2012) with I. nuttallii as a reference at 90% similarity.
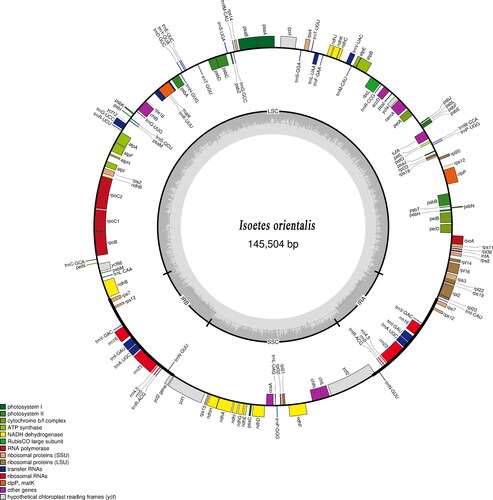
We drew the chloroplast complete genome map of this quillwort species () in OGDRAW – Draw Organelle Genome Maps (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). To find the phylogenetic position of I. orientalis, molecular phylogenetic analysis was carried out with 15 published, complete chloroplast genomes of Isoetes downloaded from GenBank.
Coding sequences (CDS) were extracted from the annotated sequences, then they were aligned using mauveAligner in Geneious Prime 2021.0.3. By employing a progressive algorithm and assuming collinearity, poorly aligned regions were excluded from the complete plastome dataset using Gblocks v0.91b in PhyloSuite v1.2.2 (Zhang et al. Citation2020). Using nucleotide as the type of sequence, up to half gap positions were allowed, and other parameters were set as default settings. For phylogenomic analysis, the resulting alignment was subjected to ML analyses performed using IQ-TREE v. 1.6.12 (Lam-Tung et al. Citation2015) with 10,000 bootstrap replicates. The best-fitting model was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017) and implemented in IQ-TREE.
Results
Complete plastid genome sequence of I. orientalis (GenBank accession: OL467336) was 145,504 bp in length containing a large single-copy (LSC) region of 91,864 bp, a small single-copy (SSC) region of 27,226 bp, and a pair of inverted repeats (IRs) of 13,207 bp each. A total of 136 genes were annotated, including 84 protein-coding genes, 37 transfer RNA (tRNA) genes, eight ribosomal RNA (rRNA) genes, and seven genes, infA, tufA, accD, rps2, rps16, a copy of ndhB and ycf2, are free in function. The overall GC content was 38.0%.
The complete plastid genome sequence of Isoetes orientalis (Isoetaceae) is 145,504 bp in length, comprising a pair of IR regions of 13,207 bp each, an LSC region of 91,864 bp, and an SSC region of 27,226 bp. The chloroplast genome contains 136 genes, including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall GC content is 38.0%.
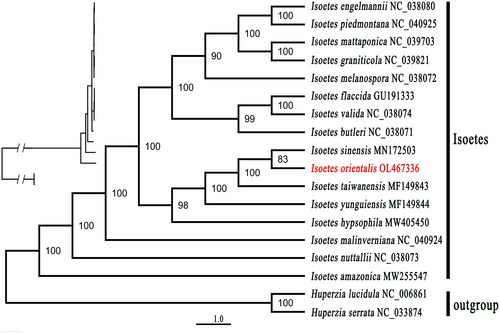
ML (maximum-likelihood) tree () indicated that Isoetes amazonica, I. nuttallii, and I. malinverniana were found as three clades solely. Isoetes orientalis was clustered with I. sinensis with support ratio 83%, while I. taiwanensis showed a sister relationship with I. sinensis and I. orientalis with 100% bootstrap support values. I. yunguienesis was a sister to the clade formed with the above three species I. taiwanensis, I. sinensis, and I. orientalis. All the above four species formed a sister group with I. hypsophila. All the five species collected from China formed sister clade with I. engelmannii, I. piedmontana, I. mattaponica, I. graniticola, I. melanospora, I. flaccida, I. valida, and I. butleri.
Discussion
Study on the evolution of Isoetaceae using complete chloroplast genomes has been carried recently (Pereira et al. Citation2021), but only a few species were employed. There are nine species of Isoetes reported in China, but only four of which were published chloroplast complete genomes. When to take further study on the Isoetaceae of China, more chloroplast complete genomes will be needed in the future. The results of the present study will supplement the chloroplast genome data of Isoetes collected in China and hold great significance in the study of this genus.
Ethical approval
The study protocol was approved by the National Orchid Conservation & Research Center of Shenzhen and South China Agricultural University. The collected plant samples did not bring any destruction to this endangered species.
Author contributions
Wrote the paper: Yanqing Li, Xi Chen, and Xiaoyan Lin; analyzed the data: Yufeng Gu and Yanqing Li. Contributed reagents/materials/analysis tools: Yuehong Yan, Baodong Liu, and Yufeng Gu. Final approval of the version to be published: Rongjing Zhang and Yufeng Gu. Funding sources: Yuehong Yan and Baodong Liu. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (148.5 KB)Acknowledgements
We thank TopEdit (www.topeditsci.com) for linguistic assistance during manuscript preparation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Genome sequence data that support the findings of this study can be obtained from GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. OL467336. Associated accession numbers are listed as BioProject PRJNA781054, SRA SRS11088792, and Bio-sample SAMN23227414.
Additional information
Funding
References
- Chen JM, Wang JY, Liu X, Zhang YW, Wang QF. 2004. RAPD analysis for genetic diversity of Isoetes sinensis. Biodivers Sci. 12(3):348–353.
- Givnish TJ, Ames M, McNeal JR, McKain MR, Steele PR, dePamphilis CW, Graham SW, Pires JC, Stevenson DW, Zomlefer WB, et al. 2010. Assembling the tree of the monocotyledons: plastome sequence phylogeny and evolution of Poales. Ann Missouri Bot Gard. 97(4):584–616.
- Givnish TJ, Spalink D, Ames M, Lyon SP, Hunter SJ, Zuluaga A, Iles WJD, Clements MA, Arroyo MTK, Leebens-Mack J, et al. 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc R Soc B. 282(1814):20151553.
- Jansen RK, Saski C, Lee SB, Hansen AK, Daniell H. 2011. Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol. 28(1):835–847.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler AV, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kang M, Ye Q, Huang H. 2005. Genetic consequence of restricted habitat and population decline in endangered Isoetes sinensis (Isoetaceae). Ann Bot. 96(7):1265–1274.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lam-Tung N, Schmidt HA, Arndt VH, Quang MB. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Liu X, Liu H, Wang QF. 2008. Spore morphology of Isoëtes (Isoëtaceae) from China. Acta Phytotaxon Sin. 446(4):479–489.
- Liu H, Wang QF, Taylor WC. 2005. Isoetes orientalis (Isoetaceae), a new hexaploid quillwort from China. Novon. 15(1):164–167.
- Lu JM, Zhang N, Du XY, Wen J, Li DZ. 2015. Chloroplast phylogenomics resolves key relationships in ferns. J Syst Evol. 53(5):448–457.
- Pang XA, Liu X, Liu H, Wu C, Wang JY, Yang SX, Wang QF. 2003. The geographic distribution and habitat of the Isoetes plants in China. Biodivers Sci. 11(4):288–294.
- Pereira JBS, Giulietti AM, Pires ES, Laux M, Watanabe MTC, Oliveira RRM, Vasconcelos S, Oliveira G. 2021. Chloroplast genomes of key species shed light on the evolution of the ancient genus Isoetes. J Syst Evol. 59(3):429–441.
- Ross TG, Barrett CF, Soto Gomez M, Lam VK, Henriquez CL, Les DH, Davis JI, Cuenca A, Petersen G, Seberg O, et al. 2016. Plastid phylogenomics and molecular evolution of Alismatales. Cladistics. 32(2):160–178.
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh GJ. 2014. From algae to angiosperms – inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 14(1):23.
- Schafran PW, Zimmer EA, Taylor WC, Musselman LJ. 2018. A whole chloroplast genome phylogeny of diploid species of Isoetes (Isoetaceae, Lycopodiophyta) in the Southeastern United States. Castanea. 83(2):224–235.
- Wei R, Yan YH, Harris AJ, Kang JS, Shen H, Xiang QP, Zhang XC. 2017. Plastid phylogenomics resolve deep relationships among eupolypod II ferns with rapid radiation and rate heterogeneity. Genome Biol Evol. 9(6):1646–1657.
- Wei R, Zhang XC. 2020. Phylogeny of Diplazium (Athyriaceae) revisited: resolving the backbone relationships based on plastid genomes and phylogenetic tree space analysis. Mol Phylogenet Evol. 143:106699.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Ye QG, Li JQ. 2003. Distribution status and causation of endangerment of Isoetes sinensis palmer in Zhejiang province. J Wuhan Bot Res. 21(3):216–220.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.