Abstract
Solanum iopetalum belongs to the Solanaceae family and is one of the tuber-bearing wild Solanum species. In this study, chloroplast genome sequencing of the species, completed with Illumina sequencing technology, is presented. The length of the chloroplast genome is 155,625 bp with a GC content of 37.86%. It comprises a large single copy (LSC) region of 86,057 bp, a small single copy (SSC) region of 18,382 bp, and two inverted repeat regions (IRa and IRb) of 25,593 bp. Additionally, 158 functional genes in the genome are identified, including 105 protein-coding genes, 8 ribosomal RNAs, and 45 transfer RNAs. Phylogenetic analysis revealed that S. iopetalum is grouped into a large clade with other Solanum species, including cultivated potatoes (S. tuberosum) and is closely related to Mexican Solanum species (S. stoloniferum, S. verrucosum, S. hougasii, S. hjertingii, and S. demissum). This study provides useful genomic information for future breeding and evolutionary studies of S. iopetalum and other Solanum species.
Introduction
Solanum iopetalum (Bitter) Hawkes 1944, which is endemic to Mexico, is one of the tuber-bearing wild relatives of cultivated potatoes (Solanum tuberosum) (Hawkes Citation1990). Due to its resistance to Phytophthora infestans (Pacheco-Sánchez et al. Citation2003; Tiwari et al. Citation2015) and its resistance to several abiotic stresses such as drought, heat, and salt (data identified in contemporary research not shown), it could be one of the great resources for potato breeding. Its endosperm balance number (EBN) of four is the same as that of S. tuberosum, which theoretically allows direct cross between the two different species for potato breeding (Hawkes Citation1990; Ortiz and Ehlenfeldt Citation1992; Cho et al. Citation1997). However, the different multiple ploidy levels causing an inconvenience in breeding must be overcome, because S. iopetalum is hexaploid and S. tuberosum is tetraploid (Hanneman Citation1994). Moreover, it has been considered an allohexaploid based on facts proposed by Hawkes (Citation1990). S. iopetalum might be produced as a hybridization of the Rotata species migrated from South America to Mexico and the Central American stellate diploids (Hawkes Citation1990). Its nuclear genome composition has been identified as allohexaploid with two (A and P) or three (A, B, and P) component genomes from diploid North American (A only, or A and B) and South American (P) species (Spooner et al. Citation2004; Pendinen et al. Citation2012). However, the results obtained from analyses of rDNA repeats and 5S rDNA suggest that S. iopetalum could be created by hybridization of closely related species, or it is autopolypoid (Volkov et al. Citation2001, Citation2003). Therefore, the origin of the Central American polyploid Solanum species S. iopetalum is still unclear, and the plastid genome of the species has rarely been studied.
Materials and methods
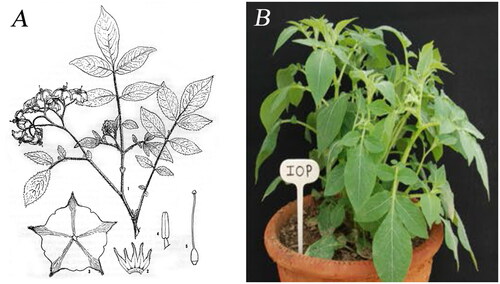
Solanum iopetalum plants (PI230459) were provided by Highland Agriculture Research Institute, South Korea (37°68′05.4″N 128°73′09.1″E) (), and the specimen was deposited in the National Agrobiodiversity Center, South Korea (http://genebank.rda.go.kr/, Young-Eun Park, [email protected]) under voucher number IT301490.
Figure 1. The species reference images for Solanum iopetalum. (A) Illustration of S. iopetalum (Correll Citation1962). (B) Plant shape of S. iopetalum (Tiwari et al. Citation2019). IOP is the three letter abbreviation for S. iopetalum.
Total genomic DNA was extracted from the leaves of the S. iopetalum plants using the Genomic DNA Extraction Kit for plants (RBC Bioscience, New Taipei City, Taiwan) according to the manufacturer’s instructions. For chloroplast genome sequencing of the species, basically performed via Phyzen bioinformatics pipeline (Kim et al. Citation2015), the library was constructed based on the PE standard protocol (Illumina, San Diego, USA). Paired-end (PE) sequencing was performed on the Illumina HiSeq2000 platform. The clean reads were assembled with the dnaLCW method using the CLC de novo assembly program in CLC assembly cell package version 4.2.1 (CLC Inc, Rarhus, Denmark). The structure and sequence of the assembled chloroplast genome were analyzed with the results of a BLASTN search at the NCBI database and BLASTZ analysis (Schwartz et al. Citation2003) using the S. gourlayi complete chloroplast genome (GenBank accession no. MH021474) as a reference. Chloroplast genome annotation was performed by using the GeSeq program (Tillich et al. Citation2017) and the circular genome map was generated using the OGDraw program (Lohse et al. Citation2007). The schematic map of the trans- and cis-splicing genes were generated using the CPGview software (Liu et al. Citation2023, http://www.1kmpg.cn/cpgview/).
Results and discussion
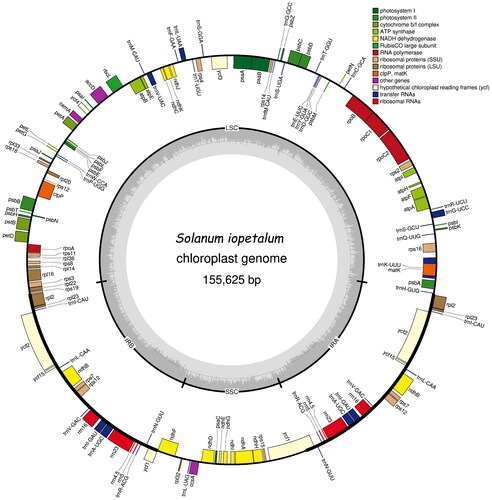
In total, approximately 2.84 Gbp of raw data were obtained and approximately 2.22 Gbp of the clean reads were assembled. The average coverage is 405.61 and mapped read depth through the whole region is approximately at least more than 200 (Supplementary Figure 1). The whole chloroplast genome sequence of S. iopetalum is 155,625 bp in length with a typical double-stranded loop structure. It is divided into four regions consisting of a large single copy (LSC) region of 86,057 bp, a small single copy (SSC) region of 18,382 bp, and two inverted repeat regions (IRa and IRb) of 25,593 bp. The overall GC content was 37.86%. The BLASTN search results showed that the sequence of S. iopetalum is most similar with that of S. gourlayi, which also originates from Mexico. The similarity between these species was also observed for the External Transcribed Spacer (ETS) sequence (Volkov et al. Citation2003) and the 5S rDNA sequence (Volkov et al. Citation2001). The S. iopetalum chloroplast genome contains a total of 158 genes with an average size of 583.1 bp, including 105 protein-coding genes, 45 transfer RNA genes, and eight ribosomal RNA genes, with average sizes of 764.7, 62.0, and 1,131.0 bp, respectively (). Eleven protein coding genes, 9 tRNA genes, and 4 rRNA genes are duplicated in the IR regions. The rps12 is a trans-splicing gene (Supplementary Figure 2A) and thirteen genes including rps16, atpF, rpoC1, ycf3, clpP, petB, petD, rpl16, rpl2, ndhB, ndhA, ndhB, and rpl2 are cis-splicing genes (Supplementary Figure 2B). Gene features are typically identical to those of higher plants. The results of chloroplast genome assembly and annotation were submitted to GenBank (http://www.ncbi.nlm.nih.gov/) under accession number MZ233587.
Figure 2. Gene map of the Solanum iopetalum chloroplast genome. Genes on the outside and inside of the map are transcribed in clockwise and counterclockwise directions, respectively.
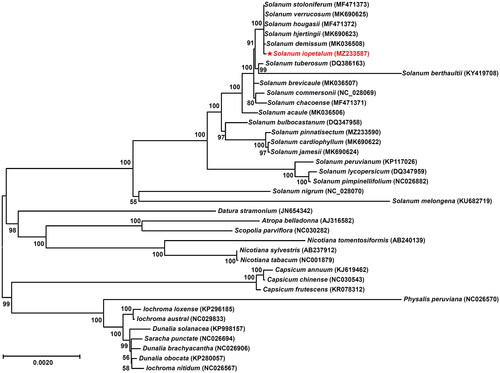
To determine the phylogenetic status of S. iopetalum, 37 other species in the Solanaceae family were obtained from the GenBank database. The phylogenetic tree was constructed using a maximum likelihood method with a Kimura 2-parameter model based on chloroplast coding sequences. The analysis was conducted using MEGA 6.0 with 1,000 bootstrapping options (Tamura et al. Citation2013). The results showed that S. iopetalum is clustered with other species in the genus Solanum and is closely related to S. stoloniferum, S. verrucosum, S. hougasii, S. hjertingii, and S. demissum in the cluster (). These five species are also Solanum members from Mexican although they have different ploidy levels and EBNs. Spooner and Sytsma (Citation1992) and Spooner et al. (Citation1995, Citation2008) reported similar results from data with chloroplast DNA restriction site variations, morphological characteristics, and cloned DNA sequences of the single-copy nuclear gene Granule-Bound Starch Synthase I (GBSSI or waxy).
Figure 3. Maximum likelihood phylogenetic tree of Solanum iopetalum with other species belonging to the Solanaceae family based on chloroplast protein coding sequences. Numbers in the nodes are the bootstrap values from 1,000 replicates. The data have been partially adopted from Park (Citation2022a). The following 38 sequences were used: MF471373 (Park Citation2018), MK690625, MF471372 (Cho et al. Citation2018), MK690623 (Park Citation2022c), MK036508 (Cho et al. Citation2019), MZ233587 (this study), DQ386163 (Gargano et al. Citation2012), KY419708 (Park Citation2017), MK036507 (Park Citation2019), NC028069 (Cho et al. Citation2016), MF471371 (Cho et al. Citation2017), MK036506 (Park Citation2021), DQ347958 (Daniell et al. Citation2006), MZ233590, MK690622, MK690624, KP117026 (Wu Citation2016), DQ347959 (Daniell et al. Citation2006), NC026882 (Wu Citation2016), NC028070 (Park Citation2016), KU682719 (Ding et al. Citation2016), JN654342, AJ316582 (Schmitz-Linneweber et al. Citation2002), NC030282 (Park and Lee Citation2016), AB240139 (Yukawa et al. Citation2006), AB237912 (Yukawa et al. Citation2006), NC001879 (Shinozaki et al. Citation1986), KJ619462 (Zeng et al. Citation2016), NC030543 (Park et al. Citation2016), KR078312 (Shim et al. Citation2016), NC026570, KP296185, NC029833, KP998157, NC026694, NC026906, KP280057, NC026567.
Conclusion
The chloroplast genome of S. iopetalum was characterized for the first time in this study. The plastome length of S. iopetalum is comparable to other Solanum species, but slightly longer than others (Park Citation2022b). Our findings will facilitate further research investigating more detailed breeding and evolutionary aspects.
Ethical approval
This study did not involve endangered or protected species, and the plant was collected with the permission from Highland Agriculture Research Institute (37°68′05.4"N 128°73′09.1"E), Pyeongchang, South Korea.
Author contributions
Tae-Ho Park did all the research, including conception and design of the work, analysis and interpretation of the data, drafting the paper and revising it critically for intellectual content, and then provided final approval of the version. The author agrees to be accountable for all aspects of the work.
Supplemental Material
Download JPEG Image (1.5 MB)Supplemental Material
Download JPEG Image (780.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available from NCBI (https://www.ncbi.nlm.nih.gov/) under accession number MZ233587. The associated BioProject, SRA, and BioSample numbers are PRJNA729825, SRR14532939, and SAMN19180224, respectively.
Additional information
Funding
References
- Cho K-S, Cheon K-S, Hong S-Y, Cho J-H, Im J-S, Mekapogu M, Yu Y-S, Park T-H. 2016. Complete chloroplast genome sequences of Solanum commersonii and its application to chloroplast genotype in somatic hybrids with Solanum tuberosum. Plant Cell Rep. 35(10):2113–2123.
- Cho K-S, Choi J-G, Cho J-H, Im J-S, Park Y-E, Hong S-Y, Park T-H. 2017. Chloroplast genome of wild tuber-bearing diploid potato relative Solanum chacoense. Mitochondrial DNA B Resour. 2(2):915–917.
- Cho K-S, Cho J-H, Im J-S, Choi J-G, Park Y-E, Jang D-C, Hong S-Y, Park T-H. 2018. The complete chloroplast genome sequence of Solanum hougasii, one of the potato wild relative species. Mitochondrial DNA B Resour. 3(2):755–757.
- Cho K-S, Cho J-H, Park Y-E, Park T-H. 2019. Chloroplast genome sequence of Solanum demissum, a wild tuber-bearing species was completed. Mitochondr DNA Part B. 4(1):1800–1802.
- Cho HM, Kim-Lee HY, Om YH, Kim JK. 1997. Influence of endosperm balance number (EBN) in interploidal and interspecific crosses between Solanum tuberosum dihaploids and wild species. Korean J Breed. 29:154–161.
- Correll DS. 1962. The potato and its wild relatives. Texas, USA: Texas Research Foundation Renner.
- Daniell H, Lee S-B, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen PK. 2006. Complete chloroplast genome sequences of Solanum bulbocatanum, Solanum lycopersicum and comparative analyses with other Solanaceae genome. Theor Appl Genet. 112(8):1503–1518.
- Ding Q-X, Liu J, Gao L-Z. 2016. The complete chloroplast genome of eggplant (Solanum melongena L.). Mitochondrial DNA B Resour. 1(1):843–844.
- Gargano D, Scotti N, Vezzi A, Bilardi A, Valle G, Grillo S, Cozzolino S, Cardi T. 2012. Genome-wide analysis of plastome sequence variation and development of plastidial CAPS markers in common potato and related Solanum species. Genet Resour Crop Evol. 59(3):419–430.
- Hanneman RE. 1994. The testing and release of transgenic potatoes in the North American Center of diversity. In: biosafety for sustainable agriculture: sharing biotechnology regulatory experience of the western hemisphere. In: Krattiger AF, Rosemarin A, editors. ISAAA: Ithaca and SEI. Stockholm; p. 47–67.
- Hawkes JG. 1990. The potato: evolution, biodiversity and genetic resources. London, UK: Belhaven Press.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resourc. 0:1–11.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- Ortiz R, Ehlenfeldt MK. 1992. The importance of endorsperm balance number in potato breeding and the evolution of tuber-bearing Solanum species. Euphytica. 60(2):105–113.
- Pacheco-Sánchez M, Lozoya-Saldaña H, Colinas-León MT. 2003. Growth regulators and cold pretreatment on in vitro androgenesis of Solanum iopetalum L. Agrociencia. 37:257–265.
- Park T-H. 2016. The complete chloroplast genome sequence of potato wild relative species, Solanum nigrum. Mitochondrial DNA B Resour. 1(1):858–859.
- Park T-H. 2017. The complete chloroplast genome of Solanum berthaultii, one of the potato wild relative species. Mitochondrial DNA B Resour. 2(1):88–89.
- Park T-H. 2018. Chloroplast genome sequence of the wild tetraploid potato relative Solanum stoloniferum. Mitochondrial DNA B Resour. 3(1):416–418.
- Park T-H. 2019. Complete chloroplast genome sequence of the wild diploid potato relative, Solanum brevicaule. Mitochondrial DNA B Resour. 4(2):4159–4160.
- Park T-H. 2021. Complete chloroplast genome sequence of the wild diploid potato relative, Solanum acaule. Mitochondrial DNA B Resour. 6(3):1189–1191.
- Park T-H. 2022a. Complete chloroplast genome sequence of Solanum hjertingii, one of the wild potato relatives. Mitochondrial DNA B Resour. 7(4):715–717.
- Park T-H. 2022b. PCR-based markers to select plastid genotypes of Solaum acaule. J Plant Biotechnol. 49(3):178–186.
- Park T-H. 2022c. Complete chloroplast genome sequence of Solanum hjertingii, one of the wild potato relative. Mitochondrial DNA B Resour. 7(4):715–717.
- Park JH, Lee J. 2016. The complete plastid genome of Scopolia parviflora (Dunn.) Nakai (Solanaceae). Korean J Plant Taxon. 46(1):60–64.
- Park HS, Lee J, Lee S-C, Yang T-J, Yoon JB. 2016. The complete chloroplast genome sequence of Capsicum chinene Jacq. (Solanaceae). Mitochondrial DNA B Resour. 1(1):164–165.
- Pendinen G, Spooner DM, Jiang J, Gavrilenko T. 2012. Genomic in situ hybridization reveals both auto- and allopolyploid origins of different North and Central American hexaploid potato (Solanum sect. Petota) species. Genome. 55(6):407–415.
- Schmitz-Linneweber C, Regel R, Du TG, Hupfer H, Herrmann RG, Maier RM. 2002. The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: the role of RNA editing in generating divergence in the process of plant speciation. Mol Biol Evol. 19(9):1602–1612.
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. 2003. Human-mouse alignments with BLASTZ. Genome Res. 13(1):103–107.
- Shim D, Raveendar S, Lee J-R, Lee G-A, Ro N-Y, Jeon Y-A, Cho G-T, Lee H-S, Ma K-H, Chung J-W. 2016. The complete chloroplast genome of Capsicum frutescens (Solanaceae). Appl Plant Sci. 4(5):1600002.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. Embo J. 5(9):2043–2049.
- Spooner DM, Berg RGvd, Rodríguez A, Bamberg J, Hijmans RJ, Cabrera SIL. 2004. Wild potatoes (Solanum section Petota: solanaceae) of North and Central America. Syst Bot Monogr. 68:1–209.
- Spooner DM, Rodríguez F, Polgár Z, Ballard HE, Jr, Jansky SH. 2008. Genomic origins of potato polyploids: GBSSI gene sequencing data. Crop Sci. 48: S27–S36.
- Spooner DM, Sytsma KJ. 1992. Reexamination of series relationships of Mexican and Central American wild potatoes (Solanum sect. Petota): evidence from chloroplast DNA restriction site variation. Syst Bot. 17(3):432–448.
- Spooner DM, van den Berg RG, Bamberg JB. 1995. Examination of species boundaries of Solanum series Demissa and potentially related species in series Acaulia and series Tuberosa (sect. Petota). Syst Bot. 20(3):295–314.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetic analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Tiwari JK, Ali N, Devi S, Zinta R, Kumar V, Chakrabarti SK. 2019. Analysis of allelic variation in wild potato (Solanum) species by simple sequence repeat (SSR) markers. 3 Biotech. 9(7):262.
- Tiwari JK, Devi S, Sharma S, Chandel P, Rawat S, Singh BP. 2015. Allele mining in Solanum germplasm: cloning and characterization of RB-homologous gene fragments from late blight resistant wild potato species. Plant Mol Biol Rep. 33(5):1584–1598.
- Volkov RA, Komarova NY, Panchuk II, Hemleben V. 2003. Molecular evolution of rDNA external transcribed spacer and phylogeny of section Petota (Genus Solanum). Mol Phylogenet Evol. 29(2):187–202.
- Volkov RA, Zanke C, Panchuk II, Hemleben V. 2001. Molecular evolution of 5S rDNA of Solanum species (section Petota): application for molecular phylogeny and breeding. Theor Appl Gent. 103:1273–1282.
- Wu Z. 2016. The completed eight chloroplast genomes of tomato from Solanum genus. Mitochondr DNA Part A. 27(6):4155–4157.
- Yukawa M, Tsudzuki T, Sugiura M. 2006. The chloroplast genome of Nicotiana sylvestris and Nicotiana tomentosiformis: complete sequencing confirms that the Nicotiana sylvestris progenitor is the material genome donor of Nicotiana tabacum. Mol Genet Genomics. 275(4):367–373.
- Zeng F, Gao C, Gao L. 2016. The complete chloroplast genome sequence of American bird pepper (Capsicum annuum var. glabriusculum). Mitochondr DNA Part A. 27(1):724–726.
