Abstract
Symbiochlorum hainandiae S.Q. Gong & Z.Y. Li, 2018 is a unicellular green alga belonging to Ulvophyceae, Chlorophyta, and plays important roles in coral reef ecosystem. In this study, high-throughput sequencing technology is used to sequence and assemble the chloroplast genome of S. hainandiae. The complete chloroplast genome of S. hainandiae was 158, 96 bp with the GC content of 32.86%. A total of 126 genes were identified, including 98 protein-coding genes, 26 tRNA, and 2 rRNA genes. The inverted repeat region was lost in the complete chloroplast genome of S. hainandiae. The phylogenetic analysis supports that S. hainandiae is a new sister lineage to the genus Ignatius within the class Ulvophyceae.
1. Introduction
Symbiochlorum hainandiae S.Q. Gong & Z.Y. Li, 2018, a unicellular green alga, belongs to Ulvophyceae, Chlorophyta, which is closely related to the genus Ignatius. The genus Symbiochlorium has been reported firstly by Gong et al. (Citation2018). So far, S. hainandiae has been only reported species within genus Symbiochlorum. Symbiochlorum hainandiae is considered as a new unidentified green algal, and isolated from bleaching coral species. Gong et al. (Citation2022) has shown that S. hainandiae is a thermotolerant alga, which can grow rapidly even at 35 °C. We also have isolated S. hainandiae in Pocillopora damicornis, and observed similar phenomenon that the alga can grow at high temperature. Therefore, we speculate that S. hainandiae may play an important role in coral reefs. However, the chloroplast genome structure of S. hainandiae and its phylogenetic position are still unclear. In this study, the first complete chloroplast genome of S. hainandiae is investigated.
2. Materials
Symbiochlorum hainandiae was collected from coral P. damicornis in the coast of Sanya City, China (18°21′ N and 109°47′ E). S. hainandiae was isolated using a single cell separation technology, and then cultured in f/2 medium with a light intensity of 80 μmol photons m−2 s−1. Symbiochlorum hainandiae was deposited at the laboratory of South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou City, Guangdong Province (http://www.scsio.ac.cn/, Fangfang Yang, [email protected]) under voucher number SY20210606.
3. Methods
The genomic DNA was extracted with the modified CTAB protocol (Doyle Citation1987). After DNA extraction, a library with the insertion size of 350 bp was constructed. Whole genome sequencing was carried out using a Novaseq 6000 sequencer (Illumina, San Diego, CA, USA). In total, the raw data totaled 4.75 G, and the clean data totaled 4.69 G after quality control processing. The coverage was showed in the Figure S1. The guanine-cytosine (GC) content of the clean data was 62.08%, the Q20 value was 97.17%, and the Q30 value was 92.50%, indicating a very high level of data quality for the complete chloroplast genome sequencing and assembly results. Cleaned reads were assembled using the de novo assembler SPAdes v.3.14.1 software. Finally, the assembled chloroplast genome was annotated using the PGA program (Qu et al. Citation2019). To understand the phylogenetic position of S. hainandiae, the complete chloroplast genomes of 19 marine microalga were selected to construct a phylogenetic tree (Turmel and Lemieux Citation2018). Multiple sequence alignment was performed by MAFFT version 7 software with the FFT-NS-2 strategy (Katoh and Standley Citation2016). The alignment results were calculated using the maximum likelihood algorithm and the phylogenetic tree was constructed by IQ-TREE 2.0 with 1000 bootstraps (Nguyen et al. Citation2015).
4. Results
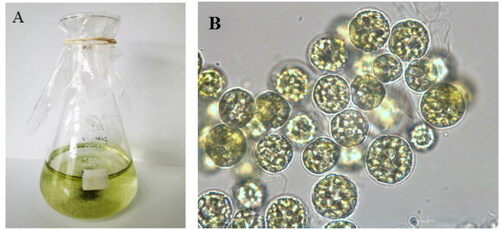
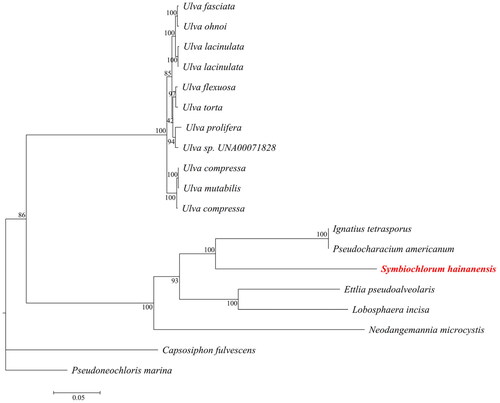
Symbiochlorum hainandiae is a type of unicellular phytoplankton, which is green, with a visible chloroplasts, starch granules, pyrenoids, and cell wall (). The complete chloroplast genome sequence of S. hainandiae was 158,961 bp in length (). This chloroplast genome did not have a quadripartite structure due to lacking large inverted repeat regions. This finds consistent with previous results (Turmel et al. Citation2017). The overall GC content was 32.86%. The chloroplast genome contained 126 genes, including 98 protein-coding genes, 26 tRNA genes, and 2 rRNA genes. The complete chloroplast genome sequence was submitted to GenBank (accession number ON645925). Phylogenetic analysis showed that S. hainandiae was a new sister lineage to the genus Ignatius within the class Ulvophyceae ().
Figure 2. Chloroplast genome map of S. hainandiae. Genes are color coded by their function in the legend. Genes on the inner and outer portions of the circle are counterclockwise directions and transcribed in clockwise, respectively.
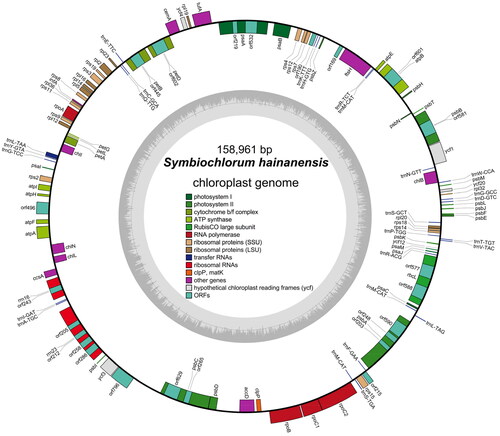
Figure 3. Maximum-likelihood phylogenetic tree based on the chloroplast genome sequences of 19 marine microalgas. The following sequences were used: Symbiochlorum hainanensis (ON645925), Pseudoneochloris marina UTEX 1445 (KY407657.1), Ulva sp. UNA00071828 (KP720616), Ulva fasciata (NC_029040.1), Ulva flexuosa (KX579943.1), Ulva torta (MZ703011.1), Ulva ohnoi (AP018696.1), Ulva compressa (MW548841.1), Ulva mutabilis (MK069584.1), Pseudoneochloris marina (KY407657.1), Lobosphaera incisa (KM821265.1), Ulva lacinulata (MW543061.1), Ulva lacinulata (NC_053614.1), Ulva prolifera (MN853879.1), Ettlia pseudoalveolaris (KM462869.1), Ignatius tetrasporus (KY407659.1), Pseudocharacium americanum (KY407658.1), Capsosiphon fulvescens (NC_039920.1), and Neodangemannia microcystis (KY407660.1).
5. Discussion and conclusion
In this study, we analyzed the chloroplast genomes information and phylogenetic relationship of S. hainandiae. We found that the inverted repeat region was lost in the complete chloroplast genome of S. hainandiae. This study provided valuable information for studying phylogenetic relationships within the class Ulvophyceae. Moreover, our study will focus on finding more algal species belonging to Symbiochlorum.
Ethical approval
The material used in this study is not designated as Endangered species. S. hainandiae is widely distributed in South China Sea. The collection of S. hainandiae was legal and reasonable.
Author contributions
Fangfang Yang was the originator of the conception and design. Yi Huang was involved with data collection and analysis. Lijuan Long designed the experiments. All authors agreed to the final version of the manuscript to be published.
Supplemental Material
Download PNG Image (40.4 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. ON645925. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA844570, SRR19521218, and SAMN28834117, respectively.
Additional information
Funding
References
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gong SQ, Jin XJ, Xiao YL, Li ZY. 2022. Ocean acidification and warming lead to increased growth and altered chloroplast morphology in the thermo-tolerant alga Symbiochlorum hainanensis. Front Plant Sci. 17:585202.
- Gong SQ, Li ZY, Zhang FL, Xiao YL, Cheng H. 2018. Symbiochlorum hainanensis gen. et sp. nov. (Ulvophyceae, Chlorophyta) isolated from bleached corals living in the South China Sea. J Phycol. 54(6):811–817.
- Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 32(13):1933–1942.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Turmel T, Otis C, Lemieux C. 2017. Divergent copies of the large inverted repeat in the chloroplast genomes of Ulvophycean green algae. Sci Rep. 7(1):994.
- Turmel T, Lemieux C. 2018. Evolution of the plastid genome in green algae. Adv Bot Res. 85:157–193.