Abstract
The complete mitochondrial genome (mitogenome) of the soft coral Sinularia acuta Manuputty and van Ofwegen, 2007 was sequenced and annotated using Illumina next-generation sequencing (NGS). The mitogenome of S. acuta was 18,730 bp in length and consisted of 14 protein-coding genes (PCGs), two ribosomal RNA genes (rRNA), and only one transfer RNA gene (tRNA-Met). The base composition was 30.18% A, 16.46% C, 19.35% G, and 34.00% T, with a total A + T content of 64.19%. The phylogenetic analysis demonstrated a close evolutionary relationship among Sinularia acuta, Sinularia penghuensis, and Sinularia maxima.
Introduction
The soft coral genus Sinularia May 1898, also called short fingered soft coral, belongs to Alcyonacea: Alcyoniidae. The species of Sinularia are distributed in extensive environmental fields in Indo-Pacific open water, from shallow to deep reefs. They have various growth patterns and different sizes of colonies (Fabricius and Alderslade Citation2001). Sinularia acuta Manuputty and van Ofwegen, 2007 was described by Manuputty and van Ofwegen from Moluccas (Indonesia) in 2007. The South China Sea is one of the habitats of this species in Southeast Asia (Benayahu et al. Citation2012, Citation2018).
The soft coral Sinularia, with more than 170 described species, is a large genus in Octocorallia; therefore, its mitochondrial genome information can draw a clear evolutionary relationship among species, but few mitochondrial genomes have been sequenced (Asem et al. Citation2019; Chen et al. Citation2019; Shen et al. Citation2021). In this study, we sequenced and annotated the complete mitochondrial genome of S. acuta (GenBank no. MW987591), and a phylogenetic analysis was performed to investigate its relationships with other Sinularia species.
Materials and methods
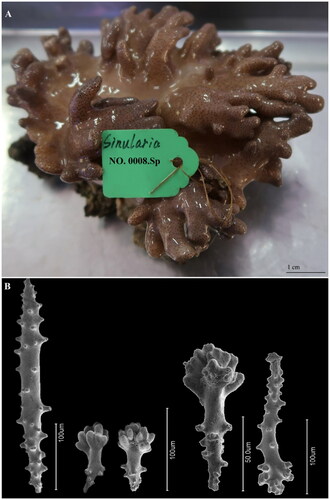
To collect the sample, permission was obtained from the Hainan Province government (Department of Science and Technology, reference number ZDYF2019154). Additionally, the specimen collection and experimental protocol of this study were approved by the Ethical Review Department of Science and Technology of Hainan Province (China) (reference number ZDKJ2019011-03-02). A live specimen of S. acuta was collected in West Island (Sanya, Hainan Province, China; 18° 14′ 5.93″ N, 109° 22′ 46.46″ E), and its taxonomic status was confirmed based on the morphology of sclerites following McFadden et al. (Citation2009). The specimen and its DNA were deposited at the Hainan Tropical Ocean University Museum of Zoology (specimen voucher number: 0008-Sp; DNA ID number: 0008-D; Chaojie Yang: [email protected]). shows the specimen reference image and morphology of sclerites.
Figure 1. (A) Image of the live individual reference specimen of Sinularia acuta Manuputty and van Ofwegen, 2007 from West Island (Sanya, Hainan Province, China; 18° 14′ 5.93″ N, 109° 22′ 46.46″ E). (B) SEM morphology of sclerites from the colony polyps and surface of S. acuta (photos by Chaojie Yanga).
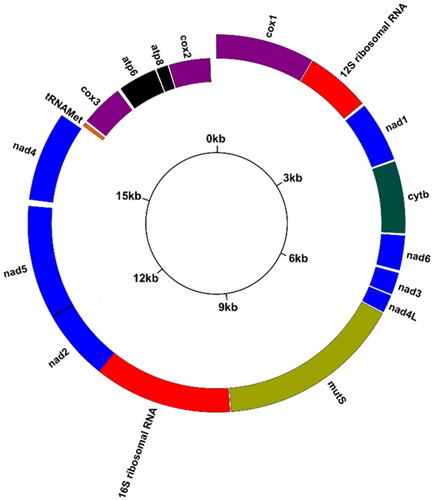
Total genomic DNA was extracted from 30 mg of tissue using Genomic Animal DNA Isolation (Sangon Biotech Co., Kit NO. B518221, Shanghai, China) (Asem et al. Citation2021). A paired-end genomic library was arranged (10 GB; two pair reads: 150 bp) utilizing the Illumina HiSeq X-ten. FastQC software was used to control the quality of the reads (Andrews Citation2010). Information on classification of raw reads and quality score distribution along reads were shown in Figure S1. The complete mitochondrial genome of Sinularia ceramensis (NC_044122) was chosen to perform the de novo assemblies by Geneious v.9.1 (Kearse et al. Citation2012). The GenomeVx online platform (http://wolfe.ucd.ie/GenomeVx/) was used to draw the mitogenomic circular map of S. acuta (). The ARWEN online platform was used to determine the position of tRNA-Met (http://130.235.46.10/ARWEN/). BioEdit software was employed to annotate the positions of protein-coding genes and ribosomal RNA genes using the reference mitogenome (Hall Citation1999). Additionally, the position and orientation of all PCGs were reconsidered by translation of amino acid sequences (ExPASy online tool: https://web.expasy.org/translate/) and the position of both the start codon and stop codon in each PCG. Maximum likelihood (ML) phylogenetic analysis was performed utilizing the software MEGA X (Kumar et al. Citation2018) with 1000 bootstrap replicates and a GTR model, which was determined by jmodeltest v.2.1.10 (Darriba et al. Citation2012). The concatenated sequences of 14 PCGs were used to draw a phylogenetic tree.
Results
Mean depth information for the assembly of complete mitochondrial genome of S. acuta was represented in Figure S2. The mitogenome of S. acuta was 18,730 bp in length. The overall base compositions were 30.18% A, 16.46% C, 19.35% G, and 34.00% T, with a total A + T content of 64.19%. Similar to the other published Alcyoniidae mitogenomes, 17 genes were detected in the mitogenome of S. acuta, including 14 PCGs, two rRNAs, and one tRNA (tRNA-Met). Ten PCGs (MutS, cox1, cytb, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6) and two rRNAs (12S and 16S) were encoded on the heavy chain. The other four PCGs (cox2, cox3, atp6, and atp8) and tRNA-Met were located on the light chain.
All PCGs used ATG as the initiation codon. Five PCGs, including nad4L, MutS, nad4, atp6, and atp8, had TAA as a stop codon, while cox1 consisted of a non-complete codon (T−). There were 16 intergenic spacers in the present mitogenome. The major non-coding region was between cox2 and cox1 (112 bp), and a 13 bp overlap was detected between nad2 and nad5.
Discussion
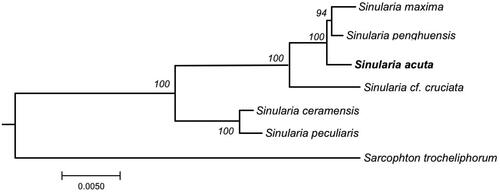
For the first time, in the present study, we reported the complete mitochondrial genome mitogenomes of Sinularia acuta which has been successfully sequenced, assembled, and annotated. The constructed phylogenetic tree revealed that Sinularia penghuensis, Sinularia maxima, and S. acuta were located in the same clade, while Sinularia ceramensis and Sinularia peculiaris were clustered together in another clade ().
Figure 3. Phylogenetic tree showing the relationship of the genus Sinularia based on the concatenated nucleotides of fourteen protein coding genes using maximum likelihood (ML). The numbers behind each node denote the bootstrap support values. The bold indicates the species: Sinularia acuta that we sequenced its mitogenome in this paper. The following sequences were used: Sinularia maxima MN485891 (Chen et al. Citation2019), Sinularia penghuensis MW256412 (Shen et al. Citation2021), Sinularia acuta MW987591 (this study), Sinularia cf. cruciata NC_034318 (Shimpi et al. Citation2017), Sinularia ceramensis NC_044122 (Asem et al. Citation2019), Sinularia peculiaris NC_018379 (Kayal et al. Citation2015), and Sarcophyton trocheliophorum MK994517 (Shen et al. Citation2019).
Conclusion
Generally, taxonomy and phylogenetic of genus Sinularia are problematic. We reported the first mitogenomes assembly and annotation of Sinularia acuta. The complete mitochondrial genome presented here could be utilized as a genomic data for further studies on phylogenetic analysis, evolutionary biology, and population genetics.
Ethical approval
The specimen collection and experimental protocol of this study were approved by the Ethical Review Department of Science and Technology of Hainan Province (China) (reference number ZDKJ2019011-03-02).
Author contributions
C.F. and C.S. designed the research. Material preparation and data collection, analyses, and interpretation were performed by C.Y. and F.M.S. The first draft of the manuscript was written by C.Y. and F.M.S. The data were reanalyzed by C.F. and C.S. The manuscript was revised by C.F. and C.S. All authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Supplemental Material
Download PDF (78.9 KB)Acknowledgments
We thank Prof. Benayahu and Prof. McFadden for their contributions to sample identification and data analysis. Our thanks to ‘editage’ (https://www.editage.cn/?utm_source=login) for English revision.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitogenome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW987591. The associated Bio-Sample, BioProject, and SRA numbers are SAMN23175316, PRJNA780668, and SRR16961931, respectively.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data [cited 2018 Oct 4]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Asem A, Eimanifar A, Li W, Shen C-Y, Shikhsarmast FM, Dan Y-T, Lu H, Zhou Y, Chen Y, Wang P-Z, et al. 2021. Reanalysis and revision of the complete mitochondrial genome of Artemia urmiana Günther, 1899 (Crustacea: Anostraca). Diversity. 13(1):14.
- Asem A, Lu H, Wang P-Z, Li W. 2019. The complete mitochondrial genome of Sinularia ceramensis Verseveldt, 1977 (Octocorallia: Alcyonacea) using next-generation sequencing. Mitochondrial DNA B Resour. 4(1):815–816. 2019.
- Benayahu Y, Ofwegen L, Dai CF, Jeng M-S, Soong K, Shlagman A, Hsieh HJ, McFadden CS. 2012. Diversity, distribution, and molecular systematics of Octocorals (Coelenterata: Anthozoa) of the Penghu Archipelago, Taiwan. Zool Stud. 51(8):1529–1548.
- Benayahu Y, van Ofwegen LP, Dai C-F, Jeng M-S, Soong K, Shlagman A, Du SW, Hong P, Imam NH, Chung A, et al. 2018. The Octocorals of Dongsha Atoll (South China Sea): an iterative approach to species identification using classical taxonomy and molecular barcodes. Zool Stud. 57:e50.
- Chen Y, Dan Y-T, Lu H, Wang P-Z, Asem A, Li W. 2019. The complete mitochondrial genome of Sinularia maxima Verseveldt (Octocorallia: Alcyonacea) using next-generation sequencing. Mitochondrial DNA B Resour. 4(2):3425–3426.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jM0delTest 2 more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Fabricius KE, Alderslade P. 2001. Soft corals and sea fans: a comprehensive guide to the tropical shallow water genera of the central-west Pacific, the Indian Ocean and the Red Sea. Townsville: Australian Institute of Marine Science.
- Hall TA. 1999. Bio Edit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 41(41):95–98.
- Kayal E, Bentlage B, Cartwright P, Yanagihara AA, Lindsay DJ, Hopcroft RR, Collins AG. 2015. Phylogenetic analysis of higher-level relationships within Hydroidolina (Cnidaria: Hydrozoa) using mitochondrial genome data and insight into their mitochondrial transcription. PeerJ. 3:e1403.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- McFadden CS, van Ofwegen LP, Beckman EJ, Benayahu Y, Alderslade P. 2009. Molecular systematics of the speciose Indo-Pacific soft coral genus, Sinularia (Anthozoa: Octocorallia). Invertebr Biol. 128(4):303–323.
- Shen C-Y, Dan Y-T, Asem A, Wang P-Z, Xue W, Tong X-B, Li W. 2019. The complete mitochondrial genome of soft coral Sarcophyton trocheliophorum (Cnidaria: Anthozoa) using next-generation sequencing. Mitochondrial DNA B Resour. 4(2):3734–3735.
- Shen C-Y, Wang P, Zh Xue W, Liu ZH, Zhao J-Y, Tong X-B, Liu CW, Wu X-F, Mao X-N, Tian S-H, et al. 2021. The complete mitochondrial genome of soft coral Sinularia penghuensis Ofwegen and Benayahu, 2012 (Octocorallia: Alcyonacea): the analysis of mitogenome organization and phylogeny. Mitochondrial DNA B Resour. 6(4):1348–1350.
- Shimpi GG, Vargas S, Poliseno A, Worheide G. 2017. Mitochondrial RNA processing in absence of tRNA punctuations in octocorals. BMC Molecular Biol. 18(1):16.