Abstract
A complete mitogenome of the Przewalski’s Wonder Gecko (Teratoscincus przewalskii) from the Junggar Basin in Northwest China was determined by using polymerase chain reaction and directly sequenced with the primer walking method. The total length was 17,184 bp, containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs) and a control region (CR). The order and structure of the genes were identical to those of congeners. The 13 PCGs contain four start codons (ATG, GTG, ATA, and ATC), three complete stop codons (TAA, TAG, and AGG), and two incomplete stop codons (T–, TA-). The concatenated PCGs were used to perform Bayesian phylogenetic analyses together with mitogenome data of the family Sphaerodactylidae and related representative taxa available in GenBank. The resulting tree recovered the monophyly of Sphaerodactylidae, and confirmed the sister relationship between T. przewalskii and T. roborowskii with strong support. The newly determined mitogenome will provide fundamental data for understanding the population genetic structure of T. pzrewalskii in particular, and the mitochondrial DNA evolution in Sphaerodactylidae in general.
Wonder geckos of the genus Teratoscincus (Squamata: Gekkota: Sphaerodactylidae) are a common component of the arid herpetofauna throughout their range from South-West Asia to Central Asia (Sindaco and Jeremcěnko Citation2008). Currently, there are nine described species of Teratoscincus across their distribution range (Uetz et al. Citation2020). Among these, the Przewalski’s Wonder Gecko (Teratoscincus przewalskii Strauch, 1887; ) is distributed in the desert regions in Northwest China and adjacent Gobi Desert in Mongolia (Zhao and Alder Citation1993; Terbish et al. Citation2006). So far, our understanding about the mitogenome evolution in Teratoscincus geckos is still limited (Macey et al. Citation2005; Li et al. Citation2016; Ma et al. Citation2020), even in the gecko family Sphaerodactylidae (Lyra et al. Citation2017; Tarroso et al. Citation2017), with the latter being the second most diverse family within Gekkota.
A complete mitochondrial genome of T. przewalskii was determined by using using Illumina Novaseq platform (Yu et al. Citation2021), which was collected from Ejin Banner, Alxa League, Inner Mongolia, China. Strikingly, in Yu et al. (Citation2021), T. przewalskii was inferred to be more closely related to T. keyserlingii than to T. roborowskii. This unexpected phylogenetic position of T. przewalskii was inconsistent with all previous findings (Macey et al. Citation1999, Citation2005; Tamar et al. Citation2021).
To test the phylogenetic hypothesis of Yu et al. (Citation2021), in this study, we report a whole mitochondrial DNA genome of T. przewalskii by Sanger sequencing, which was collected from the Junggar Basin (45.078°N, 85.391°E), Xinjiang Uygur Autonomous Region, China, in July 2017. A piece of muscle was dissected from the gecko, which had been euthanized with an overdose of sodium pentobarbital delivered by intraperitoneal injection. Both the muscle sample and the voucher specimen (LXJ0416) were fixed in 95% ethanol, and deposited at −20 °C in College of Life Science and Technology, Xinjiang University, China (contact person: Jun Li, E-mail: [email protected]). Total genomic DNA was extracted using EasyPure Genomic DNA kit (TransGen Biotech Co, Beijing, China) following the manufacturer’s instructions. Published sequences of T. keyserlingii (GenBank accession number AY753545; Macey et al. Citation2005) and T. roborowskii (GenBank accession number MT107158; Ma et al. Citation2020) were used to design 12 pairs of primers to amply and sequence T. przewalskii’s mitogenome. To obtain the sequence of the control region, we also designed some species-specific genome walking primers. Genes were annotated using the MITOS WebServer (http://mitos2.bioinf.uni-leipzig.de/index.py; Bernt et al. Citation2013) with the vertebrate genetic code.
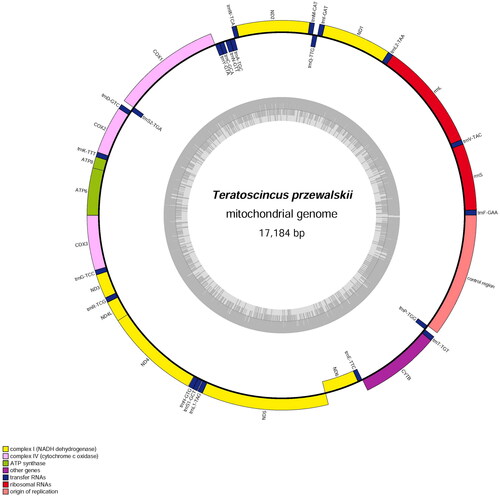
The complete mitogenome of T. przewalskii (OL471044) was a typically circular molecule with 17,184 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs) and a control region (CR) (). The gene arrangement and the composition were identical with those of congeners (Macey et al. Citation2005; Li et al. Citation2016; Ma et al. Citation2020; Yu et al. Citation2021). Except for ND6 and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser[UCA], Glu, and Pro), the remaining genes were encoded on heavy strand (H-strand). Most of the PCGs were initiated with ATN, with the exception of COX1, ATP8, ND4 and ND5 which started with GTG. ND5 was terminated with TAG; ATP8, ATP6, ND4L, ND4 and ND6 were terminated with TAA; COX1 used AGG as stop codon. The remaining PCGs (ND1, ND2, COX2, COX3, ND3 and Cytb) ended with T or TA as the incomplete stop codon. The uncorrected p-distance between our sample and that of Yu et al. (Citation2021), was 0.15%, calculated in MEGA 7 (Kumar et al. Citation2016).
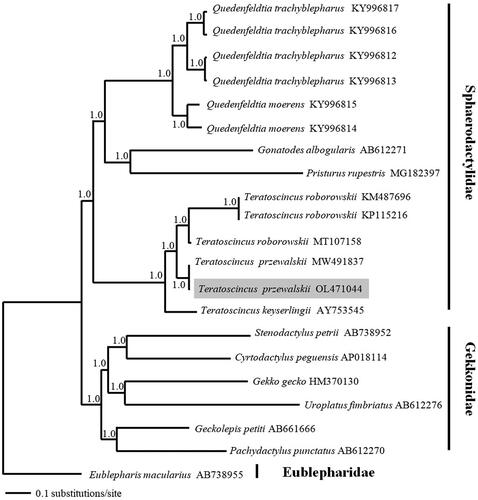
To evaluate mitochondrial sequence authenticity of T. przewalskii and its phylogenetic position, we performed phylogenetic reconstruction using mitochondrial genomes in Sphaerodactylidae available in GenBank and six species representing six genera of Gekkonidae as well as one species in Eublepharidae (Eublepharis macularius) as outgroups, with Eublepharidae as the root (Gamble et al. Citation2008). We concatenated 13 PCGs and conducted the Bayesian inference in MrBayes v.3.2 (Ronquist et al. Citation2012). The best-fitting nucleotide substitution model (GTR + G + I) for our mitochondrial dataset was selected using MrModeltest v.2.3 (Nylander, Citation2008), under the Akaike Information Criterion (AIC) (Posada and Buckley, Citation2004). As shown in , the resultant phylogenetic tree recovered the monophyly of Sphaerodactylidae and Teratoscincus (Gamble et al. Citation2008). Within the sampled Teratoscincus, T. przewalskii was recovered to be sister to T. roborowskii with strong support; which was consistent with previous findings (Macey et al. Citation1999, Citation2005; Tamar et al. Citation2021). Our results thus were contradictory to the unexpected phylogenetic findings of Ma et al. (Citation2020) and Yu et al. (Citation2021). The limited number of Teratoscincus mitogenomes used for the phylogenetic reconstructions in Ma et al. (Citation2020) and Yu et al. (Citation2021) may be the main reason which caused the differences with previous studies as well as ours. The newly determined mitogenome will provide fundamental data for understanding the population genetic structure of T. pzrewalskii in particular, and the mitochondrial DNA evolution in Sphaerodactylidae in general.
Figure 3. A 50% majority-rule consensus tree inferred from Bayesian inference using MrBayes based on the PCGs of 14 individuals of Sphaerodactylidae, 6 species in Gekkonidae as well as one species in Eublepharidae as outgroups, with Eublepharidae as the root. The novel sequencing sample is highlighted. Node numbers showed Bayesian posterior probabilities. GenBank accession numbers are given with species.
Ethical approval
Ethical approval for the study was obtained from the Ethical Committee of College of Life Science and Technology, Xinjiang University.
Author contributions
Jun Li designed the study, collected specimen in the field and performed molecular experiments. Jun Li and Xianguang Guo performed data analysis, drafted the manuscript. Both authors have read and agreed to be accountable for all aspects of this work.
Supplemental Material
Download MS Word (13.7 KB)Acknowledgements
We thank Yongfei He for his contribution to sample collecting in the fieldwork.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that supports the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, with reference number OL471044.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Gamble T, Bauer AM, Greenbaum E, Jackman TR. 2008. Evidence for Gondwanan vicariance in an ancient clade of gecko lizards. J Biogeogr. 35:88–104.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li HM, She Y, Hou LX, Zhang Y, Guo DN, Qin XM. 2016. The complete mitochondrial genome of Teratoscincus roborowskii (Squamata: Gekkonidae). Mitochondrial DNA A. 27(3):1916–1917.
- Lyra ML, Joger U, Schulte U, Slimani T, El Mouden EH, Bouazza A, Künzel S, Lemmon AR, Lemmon EM, Vences M. 2017. The mitochondrial genomes of Atlas Geckos (Quedenfeldtia): mitogenome assembly from transcriptomes and anchored hybrid enrichment datasets. Mitochondrial DNA B Resour. 2(1):356–358.
- Ma JL, Dai XY, Xu XD, Guan JY, Zhang YP, Zhang Zhang JY, Yu DN. 2020. The complete mitochondrial genome of Teratoscincus roborowskii (Squamata: Gekkonidae) and its phylogeny. Mitochondrial DNA B. 5(2):1575–1577.
- Macey JR, Fong JJ, Kuehl JV, Shafiei S, Ananjeva NB, Papenfuss TJ, Boore JL. 2005. The complete mitochondrial genome of a gecko and the phylogenetic position of the Middle Eastern Teratoscincus keyserlingii. Mol Phylogenet Evol. 36(1):188–193.
- Macey JR, Wang YZ, Ananjeva NB, Larson A, Papenfuss TJ. 1999. Vicariant patterns of fragmentation among gekkonid lizards of the genus Teratoscincus produced by the Indian collision: a molecular phylogenetic perspective and an area cladogram for Central Asia. Mol Phylogenet Evol. 12(3):320–332.
- Nylander JAA. 2008. MrModeltest 2.3. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53(5):793–808.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sindaco R, Jeremcěnko VK. 2008. The reptiles of the Western Palearctic. 1. Annotated checklist and distributional atlas of the turtles, crocodiles, amphisbaenians and lizards of Europe, North Africa, Middle East and Central Asia. Latina: Edizioni Belvedere.
- Tamar K, Els J, Kornilios P, Soorae P, Tarroso P, Thanou E, Pereira J, Shah JN, Elhassan EEM, Aguhob JC, et al. 2021. The demise of a wonder: evolutionary history and conservation assessments of the Wonder Gecko Teratoscincus keyserlingii (Gekkota, Sphaerodactylidae) in Arabia. PLoS ONE. 16(1):e0244150.
- Tarroso P, Simo-Riudalbas M, Carranza S. 2017. The complete mitochondrial genome of Pristurus rupestris rupestris. Mitochondrial DNA B. 2(2):803–803.
- Terbish K, Munkhbayar K, Clark EL, Munkhbat J, Monks EM, Munkhbaatar M, Baillie JEM, Borkin L, Batsaikhan N, Samiya R, et al. 2006. Mongolian red list of reptiles and amphibians. Regional red list series. Vol. 5. London: Zoological Society of London.
- Uetz P, Freed P, Hošek J, editors. 2020. The reptile database. [accessed 2021 March 23]. http://www.reptile-database.org
- Yu H, Liu Y, Liu Y, Yang J, Li S, Bi J, Zhang R. 2021. Complete mitochondrial genome of Teratoscincus przewalskii (Reptilia, Squamata, Sphaerodactylidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 6(11):3166–3168.
- Zhao E-M, Alder K. 1993. Herpetology of China. Oxford: Society for the Study of Amphibians and Reptiles.