Abstract
The complete mitochondrial genome of Trematomus newnesi was sequenced using an Illumina platform. The 18,602 bp mitogenome contains 13 protein-coding genes, two rRNAs, and 23 tRNAs (tRNAMet is duplicated). The eight stop codons are TAA, TAG, CTT, GTA, AAT, ACT, AGG, and TTA. Two start codons ATG and GTG are present. The GC content is 44.4% and AT content is 55.6%. A phylogenetic tree was generated using 13 species from three families. The results showed that T. newnesi is closely related to Pagothenia borchgrevinki in Nototheniidae. This study provides fundamental data for further genetic evolutionary studies on T. newnesi.
Introduction
The suborder Notothenioidei is the dominant fish fauna distributed in the Antarctic Ocean. This suborder accounts for almost all (approximately 95%) of the total fish biomass in this polar region (Eastman Citation2005). Notothenioids are characterized by the presence of antifreeze glycoproteins (AFGPs), lack of hemoglobin, and/or heat-shock response capacity in some species (DeVries and Wohlschlag Citation1969; Hofmann et al. Citation2000; Buckley et al. Citation2004; Beers and Jayasundara Citation2015; Kim et al. Citation2019). Dusky rockcod, Trematomus newnesi (Boulenger Citation1902) is a demersal fish that is distributed in depths ranging from 0 to 400 m, often found in shallow near shore areas and intertidal zones according to the fish base (https://www.fishbase.se/summary/7058) (Miller Citation1993) (). The genus Trematomus has a total of 11 species. The whole mitochondrial genome sequence has been reported from only five species (Song et al. Citation2016b; Alam et al. Citation2019; Choi et al. Citation2021; Papetti et al. Citation2021). In this study, we provide a complete mitochondrial genome of T. newnesi with the phylogenetic analysis to increase the understanding of its evolution compared to other Antarctic fishes.
Materials
The fish were collected near the Jang Bogo Station (Korean Antarctic Research Base) in the north of Victoria Land (74°37.8792′S, 164°14.8883′E) from December 2020 to February 2021, stored at −20 °C immediately after death, and transferred to the Korea Polar Research Institute (KOPRI), Incheon, Korea. A specimen was deposited at KOPRI (https://www.kopri.re.kr/eng/, Jin-Hyoung Kim, [email protected]) under voucher number (Antarctic fish_012).
Methods
The total genomic DNA was extracted from the muscle tissue using the conventional phenol-chloroform method. An Illumina paired-end library was prepared from the extracted DNA following the manufacturer’s instructions. Sequencing was performed on an Illumina HiSeq XTM Ten by a commercial company (Phyzen, Seong-Nam, South Korea). Adapter sequences and low-quality reads were eliminated using quality_trim option under Phred score 20; the pre-processing completed data was performed de novo assembly using QIAGEN CLC Assembly Cell package (Version 4.21, CLC Inc., Aarhus, Denmark). Subsequently, contigs derived from the mitochondrial genome were selected, and the final mitochondrial genome was assembled using Illumina data-based polishing. Next, the complete mitochondrial genome sequence was annotated using the GeSeq program (https://chlorobox.mpimp-golm.mpg.de/geseq-app.html) based on related species. Manual curation was performed using the Artemis annotation tool (Liang et al. Citation2018) with NCBI BLASTN search.
The phylogenetic analysis was performed by MEGA11 (Lin et al. Citation2012) involved 13 protein-coding genes. The following sequences were used: KT921282.1 (Song et al. Citation2016a), NC039543.1 (Liang et al. Citation2018), JF933907.1, GU217678.1 (Lin et al. Citation2012), JF933906.1, AB723627.1, JF933905.1, KU951144.1 (Liu et al. Citation2016), MT447073.1 (Choi et al. Citation2021), NC053364.1 (Song et al. Citation2016b), NC057674.1 (Papetti et al. Citation2021), NC057675.1 (Papetti et al. Citation2021). The Maximum Likelihood (ML) tree was constructed with 1000 bootstrap replicates and JTT matrix-based model (Felsenstein Citation1981).
Results and discussions
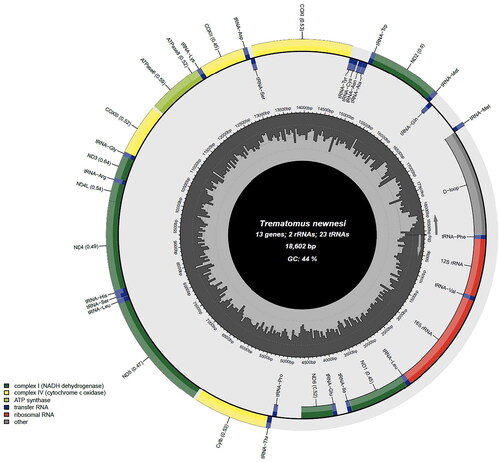
The complete mitochondrial genome of T. newnesi (GenBank Number: OP432732.1) is 18,602 bp in length and consists of 13 protein-coding genes (ND1, ND2, ND3, ND4, ND4L, ND5, ND6, CYTB, COX1, COX2, COX3, ATP6, ATP8), two rRNA genes (rrnL and rrnS), and 23 tRNA genes (tRNAMet was duplicated) (). In the 23 tRNA genes, the duplication of tRNAMet gene might because T. newnesi have a high frequency of gene duplications and rearrangement of mitochondrial genomes (Lin et al. Citation2012; Song et al. Citation2016b; Papetti et al. Citation2021; Minhas et al. Citation2022; Patel et al. Citation2022). In 13 protein-coding genes, GTG was the start codon for COX1, and ATG was the start codon for the rest. There were eight stop codons, TAA for COX1, ATP6, CYTB, ND4L, and ND1 genes, and the stop codon TAA of five genes (COX1, ATP6, CYTB, ND4L, and ND1) was predicted to be completed by the addition of 3′ A residues to the mRNA, TAG for ATP8 and ND5 genes, CTT for COX2 gene, GTA for COX3 gene, AAT for ND3 gene, ACT for ND4 gene, AGG for ND6 gene, and TTA for ND2 gene. The GC contents were 44.4% and AT contents were 55.6%. These results are corresponded with a previous result regarding of the inversion of the control region and mechanisms of rearrangement in notothenioid mitochondrial genomes (Papetti et al. Citation2021). In particular, it is confirmed that the mitochondrial genome of T. newnesi also has an extremely rare inversion of a large genomic segment as shown in other fishes of Trematominae such as Trematomus eulepidotus, Trematomus tokarevi, Trematomus borchgrevinki, Lindbergichthys nudifrons (Papetti et al. Citation2021).
Figure 2. Circular gene map of the Trematomus newnesi mitochondrial genome using MitoFish (http://mitofish.aori.u-tokyo.ac.jp/annotation/input/).
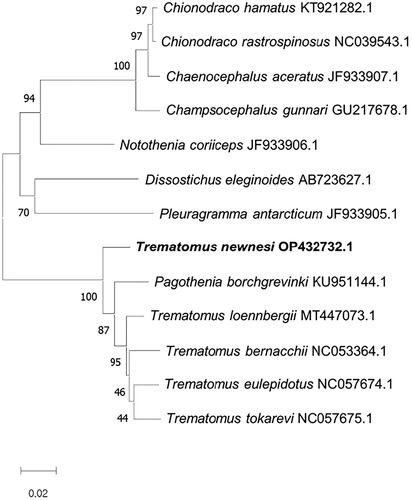
The phylogenetic tree () showed that T. newnesi was close to Pagothenia borchgrevinki in Nototheniidae. The phylogenetic analysis would provide an intuitive insight into the evolutionary relationship of T. newnesi with other fish species living in the Antarctic Ocean.
Author contributions
Conceptualization, JHK, PTN, SL, and JK; methodology, PTN, SL, JJ, JK, and DWH; resources, DWH, ICK, and JP; writing original draft preparation, PTN, SL, and JJ; writing review and editing JHK, ICK, JHL, and JP; interpretation of data, JHK, PTN, SL, ICK, JP, and JHL; project administration, JHK; funding acquisition, JHL and JHK. All the authors have read and agreed to the published version of the manuscript.
Ethics statement
All handling and experimental procedures followed the ethical guidelines regulated by the Animal Experimental Ethics Committee established by the Korea Polar Research Institute (KACC2201-007).
Supplemental Material
Download JPEG Image (51.7 KB)Acknowledgments
We thank PHYZEN Co. for their assistance with software application and analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are available in GenBank at https://www.ncbi.nlm.nih.gov/under accession no. OP432732.1. The associated BioProject, SRA, and Bio-Sample numbers were PRJNA881762, SRR21669782, and SAMN30864146, respectively.
Additional information
Funding
References
- Alam MJ, Kim J-H, Andriyono S, Lee J-H, Lee SR, Park H, Kim H-W. 2019. Characterization of complete mitochondrial genome and gene organization of sharp-spined notothenia, Trematomus pennellii (Perciformes: Nototheniidae). Mitochondrial DNA Part B. 4(1):648–649.
- Beers JM, Jayasundara N. 2015. Antarctic notothenioid fish: what are the future consequences of ‘losses’ and ‘gains’ acquired during long-term evolution at cold and stable temperatures? J Exp Biol. 218(12):1834–1845.
- Boulenger GA. 1902. Pisces. Pt 5: 174–189, Pls. 11–18. In: Lancester ER, editor. Report on the collections of natural history made in the Antarctic regions during the voyage of the “Southern Cross”. British Museum (Natural History), London: ix + 344 pp., 53 leaves of plates. 1.
- Buckley BA, Place SP, Hofmann GE. 2004. Regulation of heat shock genes in isolated hepatocytes from an Antarctic fish, Trematomus bernacchii. J Exp Biol. 207(Pt 21):3649–3656.
- Choi E, Im T-E, Lee SJ, Jo E, Kim J, Kim SH, Chi YM, Kim J-H, Park H. 2021. The complete mitochondrial genome of Trematomus loennbergii (Perciformes, Nototheniidae). Mitochondrial DNA Part B. 6(3):1032–1033.
- DeVries AL, Wohlschlag DE. 1969. Freezing resistance in some Antarctic fishes. Science. 163(3871):1073–1075.
- Eastman JT. 2005. The nature of the diversity of Antarctic fishes. Polar Biol. 28(2):93–107.
- Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 17(6):368–376.
- Hofmann GE, Buckley BA, Airaksinen S, Keen JE, Somero GN. 2000. Heat-shock protein expression is absent in the Antarctic fish Trematomus bernacchii (Family Nototheniidae). J Exp Biol. 203(15):2331–2339.
- Kim B-M, Amores A, Kang S, Ahn D-H, Kim J-H, Kim I-C, Lee JH, Lee SG, Lee H, Lee J, et al. 2019. Antarctic blackfin icefish genome reveals adaptations to extreme environments. Nat Ecol Evol. 3(3):469–478.
- Liang S, Song W, Huang H, Qu T, Zhang F, Jiang K, Chen X, Ma L. 2018. The complete mitochondrial genome of Chionodraco rastrospinosus (Notothenioidei: Channichthyidae) with phylogenetic consideration. Mitochondrial DNA Part B. 3(2):816–817.
- Lin C-Y, Lin W-W, Kao H-W. 2012. The complete mitochondrial genome of the mackerel icefish, Champsocephalus gunnari (Actinopterygii: Channichthyidae), with reference to the evolution of mitochondrial genomes in Antarctic notothenioids. Zool J Linn Soc. 165(3):521–533.
- Liu Y, Yang M, Zhou T, Xing H, Chen L, Zhang D. 2016. Complete mitochondrial genome of the Antarctic cod icefish, Pagothenia borchgrevinki (Perciformes: Nototheniidae). Mitochondrial DNA Part B. 1(1):432–433.
- Miller RG. 1993. History and atlas of the fishes of the Antarctic Ocean. Foresta Institute for Ocean and Mountain Studies.
- Minhas BF, Beck EA, Cheng CC, Catchen J. 2022. Novel mitochondrial genome rearrangements including duplications and extensive heteroplasmy in Antarctic notothenioid fishes. BioRxiv. DOI:10.1101/2022.09.19.508608.
- Papetti C, Babbucci M, Dettai A, Basso A, Lucassen M, Harms L, Bonillo C, Heindler FM, Patarnello T, Negrisolo E, et al. 2021. Not frozen in the ice: large and dynamic rearrangements in the mitochondrial genomes of the Antarctic Fish. Genome Biol Evol. 13(3):evab017.
- Patel S, Evans CW, Stuckey A, Matzke NJ, Millar CD. 2022. A unique mitochondrial gene block inversion in Antarctic Trematomin fishes: a cautionary tale. J Hered. 113(4):414–420.
- Song W, Li L, Huang H, Meng Y, Jiang K, Zhang F, Chen X, Ma L. 2016a. The complete mitochondrial genome of Chionodraco hamatus (Notothenioidei: Channichthyidae) with phylogenetic consideration. Mitochondrial DNA Part B. 1(1):52–53.
- Song W, Li L, Huang H, Zhao M, Jiang K, Zhang F, Zhao M, Chen X, Ma L. 2016b. The complete mitochondrial genome sequence and gene organization of Trematomus bernacchii (Perciformes: Nototheniidae) with phylogenetic consideration. Mitochondrial DNA Part B. 1(1):50–51.