Abstract
For the first time, this study presents the complete mitochondrial genome and phylogenetic analysis of Eonemachilus longidorsalis, an endemic species in southwest China. The mitogenome is a circular molecule of 16,569 bp in length with a base composition of 30.2% A, 27.3% T, 16.6% G, and 25.9% C. The phylogenetic analysis based on the complete mitogenomes shows that E. Longidorsalis clusters with Yunnanilus jiuchiensis, Yunnanilus pleurotaenia and Eonemachilus niger. This contribution may provide a valuable framework for completely resolving phylogenetic relationships of the family Nemacheilidae in future research.
Introduction
Eonemachilus longidorsalis (Li, Tao and Lu 2000) is distributed in the eastern Yunnan, Nanpangjiang drainage of China. This species combines morphological characteristics that can be distinguished from other Eonemachilus fishes: scales covering the whole body; extremely long dorsal fin iii-11-12; anal fin ii-7; pectoral fin i-10; caudal fin i-14-i; 12 inner gill rakers on the first gill-arch and no outer gill rakes (Du et al. Citation2021). Eonemachilus (Berg Citation1938) was once regarded as a synonym of Yunnanilus by some ichthyologists (Kottelat and Chu Citation1988; Zhu Citation1989; Yang Citation1991), but morphological studies supported that Eonemachilus was a valid genus, which can be distinguished from Yunnanilus based on absent (vs. present) lateral line and cephalic lateral-line canals (Li WX et al. Citation2000; Kottelat Citation2012). This viewpoint was confirmed by the evolutionary position of E. nigomaculatus based on molecular data, which was the most basal clade of the tribes Lefuini, Nemacheilini, Triplophysini, and some members of Yunnanilini (Du et al. Citation2021). However, we noticed that the sequence of E. nigomaculatus was misidentified and changed in GenBank by the author, resulting in the invalidity of previous phylogenetic analyses conducted from molecular perspective. In this study, the complete mitochondrial genome of E. longidorsalis was determined and described, and its taxonomic status in Nemacheilidae fishes was analyzed, which may provide a valuable genetic resource for further phylogeny analysis on the genus Eonemachilus.
Materials
The E. longidorsalis specimen was collected from Nanpangjiang, Qujing, Yunnan Province of China (25°58′76.38″N, 103°83′05.79″E), and it was identified according to morphological keys () (Du et al. Citation2021). Then the sample was preserved in 95% ethanol and deposited in Aquatic Science and Technology Institution Herbarium under the voucher number ASTIH-21b1108d27 (https://www.jsahvc.edu.cn/; person in charge of the collection: Lin Song; email: [email protected]).
Methods
After being transported to Shanghai Genesky Biotechnologies Inc, total genomic DNA was extracted from muscle using the Tguide Cell/tissue genomic DNA Extraction Kit (OSR-M401) (Tiangen, Beijing, China). The next several steps were DNA sample quality control, DNA library construction, PCR amplification, size selection, library quality check, and library pooling. Sequencing was conducted on the Illumina HiSeq 4000 Sequencing platform (Illumina, CA, USA). The complete mitogenome of E. longidorsalis was obtained by sequence assembly on MetaSPAdes software (Nurk et al. Citation2017). The annotation process was executed using MitoMaker (Bernt et al. Citation2013). The mitogenome sequence and gene annotations were submitted to the National Center for Biotechnology Information GenBank database under the accession number OM732331. The CGView online server (https://proksee.ca/) was chosen to draw the mitogenome map.
To infer the taxonomic status of E. longidorsalis, the complete mitochondrial genomes of 31 Nemacheilidae species and 2 Balitoridae species were concatenated into a dataset, and a phylogenetic tree was constructed via the maximum likelihood method under the most suitable nucleotide sequence model GTR + G+I on MEGA X (Kumar et al. Citation2018). The 2 Balitoridae species, Sinogastromyzon szechuanensis (Li W et al. Citation2021) and Metahomaloptera omeiensis (Li Y et al. Citation2016) were used as outgroups.
Results
Mitogenome organization
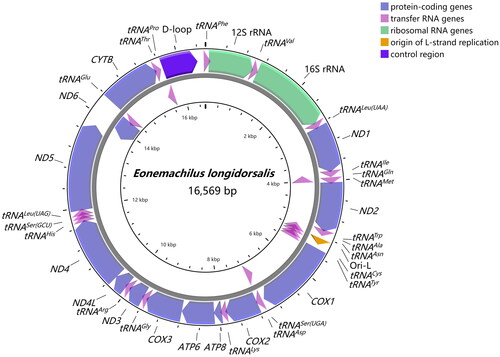
The circular mitogenome is 16,569 bp in length, encompassing 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and a control region (D-loop) (). The nucleotide composition of E. longidorsalis appears to be 30.2% A, 27.3% T, 16.6% G, and 25.9% C, with an AT bias of 57.5%. Most genes are encoded on the H-strand except for ND6 and 8 tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu, and tRNAPro). 22 transfer RNA genes range from 67 to 76 bp in length. The large ribosomal gene (16S) located between tRNAVal and tRNALeu(UUR) is 1,657 bp long, and the small (12S) located between tRNAPhe and tRNAVal is 953 bp long. Furthermore, the noncoding control region located between tRNAPhe and tRNAPro is 745 bp in length. Among the 13 protein-coding genes, the lengths vary from 168 bp (ATP8) to 1,839 bp (ND5). All PCGs conventionally utilize ATG as the start codon with the exception of COX1, which starts with GTG. 3 PCGs (COX2, ND4, and CYTB) end with a single nucleotide T, and COX3 sets incomplete TA as the stop codon. 6 PCGs (ND1, COX1, ATP8, ATP6, ND4L, and ND5) terminate with canonical TAA, while 3 PCGs (ND2, ND3, and ND6) end with complete TAG. Sequence analyses display that the mitogenome has 2 overlaps between tRNAs (tRNAIle-tRNAGln, tRNAThr-tRNAPro), and 4 overlaps between protein-coding genes (ATP8-ATP6, ATP6-COX3, ND4L-ND4, ND5-ND6).
Figure 2. Gene map of the mitochondrial genome of Eonemachilus longidorsalis. Gene encoded on H- and L- strands with inverse arrow directions were shown outside and inside the circle, respectively. The complete mitogenome of E. longidorsalis is 16,569 bp with the inclusion of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, origin of L-strand replication (Ori-L) and control region (D-loop).
Phylogenetic analysis
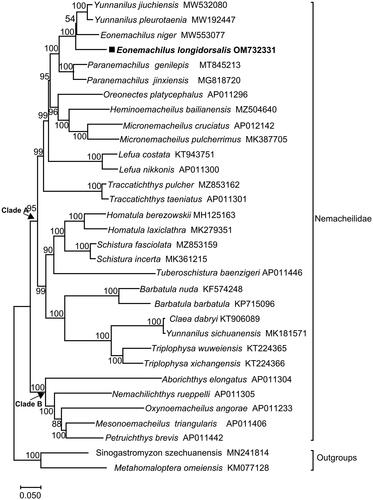
As shown in , based on the complete mt DNA genome sequences of 33 species, the family Nemacheilidae is divided into two clades: 14 genera (Yunnanilus, Eonemachilus, Paranemachilus, Oreonectes, Heminoemacheilus, Micronemacheilus, Lefua, Traccatichthys, Homatula, Schistura, Tuberoschistura, Barbatula, Claea and Triplophysa) form Clade A, while 5 genera (Aborichthys, Nemachilichthys, Oxynoemacheilus, Mesonoemacheilus and Petruichthys) form Clade B. Meanwhile, E. longidorsalis is clustered with Yunnanilus jiuchiensis (Du et al. Citation2021), Yunnanilus pleurotaenia and Eonemachilus niger. Notably, Yunnanilus sichuanensis is clustered with Claea dabryi, which indicates that the genus Yunnanilus is complexible.
Figure 3. Maximum-likelihood (ML) phylogenetic tree reconstructed using concatenated mitochondrial genomes of E. longidorsalis and other 32 fishes. Accession numbers are indicated after the species names. The tree topology was evaluated by 1000 bootstrap replicates. Bootstrap values at the nodes correspond to the support values for ML methods.
Discussion and conclusion
Through next-generation sequencing and assembly, the complete mitogenome of E. longidorsalis is 16,569 bp in length. The gene order and composition are identical to typical mitogenomes of other teleost fishes (Luo et al. Citation2019). The maximum likelihood tree based on the complete mitochondrial genomes of E. longidorsalis and 32 other species supports that E. longidorsalis constitutes a sister-group mitogenome relationship with Yunnanilus jiuchiensis, Yunnanilus pleurotaenia and Eonemachilus niger. The inclusion of more related taxa on future whole mitogenome phylogenetic analyses may help to understand the phylogeny of Yunnanilus and Eonemachilus (Yamasaki et al. Citation2015; Wang et al. Citation2019; Maeda et al. Citation2021). In conclusion, this study will provide important information for future taxonomic, systematic, and genetic studies of Nemacheilidae.
Ethical approval
Experiments were performed following the recommendations of the Ethics Committee for Animal Experiments of Jiangsu Agri-animal Husbandry Vocational College. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Authors’ contributions
LS and X-JC: conceptualization and workflow design; QW and QW: data acquisition and analysis. All authors were involved in drafting the paper and final version approval. The contributions are ranked in order.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the reference number OM732331. The associated ‘BioProject,’ ‘Bio-Sample’ and ‘SRA’ numbers are PRJNA816066, SAMN26656524, and SRR18326284 respectively.
Additional information
Funding
References
- Berg LS. 1938. On some South China loaches (Cobitidae, Pisces). Bulletin de la Société Impériale Des Naturalistes de Moscou. 47:314–318.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Du LN, Yang J, Min R, Chen XY, Yang JX. 2021. A review of the cypriniform tribe Yunnanilini prokofiev, 2010 from China, with an emphasis on five genera based on morphologies and complete mitochondrial genomes of some species. Zool Res. 42(3):310–334.
- Kottelat M. 2012. Conspectus cobitidum: an inventory of the loaches of the world (Teleostei: Cypriniformes: Cobitoidei). The Raffles Bulletin of Zoology. S26:1–199.
- Kottelat M, Chu XL. 1988. Revision of Yunnanilus with descriptions of a miniature species flock and six new species from China (Cypriniformes: Homalopteridae). Environ Biol Fishes. 23(1–2):65–94.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li W, Chen X, Zheng M, Wang Y, Zhou X, Li H, Zhou G, Li B. 2021. Complete mitochondrial genome of Sinogastromyzon szechuanensis (Teleostei, Cypriniformes, Balitoridae) obtained using next-generation sequencing. Mitochondrial DNA B Resour. 6(1):69–70.
- Li WX, Tao JN, Mao WN, Lu ZM. 2000. Two new species of Yunnanilus from eastern Yunnan, China (Cypriniformes: Cobitidae). Aata Zootaxonomica Sinica (in Chinese.). 25(3):349–353.
- Li Y, Tang M, Xue Y, Chen HJ, Ye Q, Li Y. 2016. Complete mitochondrial genome of the Chinese balitorine loach, Metahomaloptera omeiensis (Teleostei, Cypriniformes). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1449–1450.
- Luo F, Huang J, Luo T, Yang J, Wen Y. 2019. Complete mitochondrial genome and phylogenetic analysis of Yunnanilus pulcherrimus (Cypriniformes, Nemacheilidae). Mitochondrial DNA B. 4(1):1269–1270.
- Maeda K, Shinzato C, Koyanagi R, Kunishima T, Kobayashi H, Satoh N, Palla HP. 2021. Two new species of Rhinogobius (Gobiiformes: Oxudercidae) from Palawan, Philippines, with their phylogenetic placement. Zootaxa. 5068(1):81–98.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Wang D, Dai CX, Li Q, Li Y, Liu ZZ. 2019. Complete mitochondrial genome and phylogenic analysis of Rhinogobius cliffordpopei (Perciformes, Gobiidae). Mitochondrial DNA B Resour. 4(2):2473–2474.
- Yamasaki YY, Nishida M, Suzuki T, Mukai T, Watanabe K. 2015. Phylogeny, hybridization, and life history evolution of Rhinogobius gobies in Japan, inferred from multiple nuclear gene sequences. Mol Phylogenet Evol. 90:20–33.
- Yang JX. 1991. The fishes of Fuxian Lake, Yunnan, China, with description of two new species. Ichthyological Exploration of Freshwaters. 2(3):193–202.
- Zhu SQ. 1989. The loaches of the subfamily Nemacheilinae in China. Nanjing (China): Jiangsu Science and Technology Publishing House (in Chinese); p. 14–20.

