Abstract
Pseudostellaria davidii (Franch.) Pax belongs to subseries distancs of Pseudostellaria (Caryophyllaceae), and is mainly distributed in north-eastern Asia. The complete chloroplast (cp) genome of P. davidii was assembled and annotated for the first time in this study. The cp genome of P. davidii is 149,732 bp in length with the GC content of 36.57%, and it consists of four subregions: a large single-copy (LSC) region of 81,156 bp, a small single-copy (SSC) region of 16,894 bp and two inverted repeats (IR) regions of 25,841 bp each. The cp genome of P. davidii encodes a total of 111 unique genes, which are 77 protein-coding genes, four rRNA genes, and 30 tRNA genes. The results of phylogenetic analysis strongly suggested that Pseudostellaria was a monophyletic group and P. davidii forms an independent sister clade to other species of Pseudostellaria.
Introduction
Pseudostellaria davidii was first established by Pax in 1934 as a new species of the tribe Alsineae in Caryophyllaceae and mainly distributed in north-eastern Asia (Pax et al. Citation1934; Zeng et al. Citation2016). P. davidii was often misidentified due to having minimal external characters to species such as P. palibiniana and P. japonica. Among Pseudostellaria, P. davidii has five petals and sepals,10 stamens, two or three styles, and one or two short napiform roots (). The root shape and distribution of sepal hairs can be used as diagnostic characters to distinguish P. davidii and P. palibiniana. The pollen of P. davidii has spheroidal grains with 25.76 μm in diameter and 15 round pores with a distance of 6.28 μm apart from each other, which is larger than that of P. japonica (Cui et al. Citation2020). In addition, a natural hybrid between P. davidii and P. palibiniana has been verified through the analysis of morphological characters, somatic chromosome numbers, pollen sterility, and random amplified polymorphic DNA (RAPD) (Choi et al. Citation2001). Due to containing valuable information with a highly conservative nature, the complete chloroplast (cp) genome has been widely used in molecular markers, barcode identification, phylogenetic analysis and other fields (Yang et al. Citation2020; Gu et al. Citation2022). Therefore, we characterized the structure of the cp genome of P. davidii and analyzed its phylogenetic relationship with other species in Caryophyllaceae family.
Materials and methods
Fresh leaves of Pseudostellaria davidii were collected from Tonghua City, Jilin Province (N41°43', E125°56'). The voucher specimen was stored in the herbarium at the Chengde Medical University (http://www.cdmc.edu.cn/, Jinxin Liu, [email protected]), voucher number is HPAA0142. Total genomic DNA was extracted from P. davidii fresh leaves by using the universal genomic DNA extraction kit (Fansheng TCM Technology Co., Ltd, China). The concentration and quality of the extracted DNA were then determined using Qubit 4.0 (Thermo Fisher Scientific, Inc., USA). Genomic DNA was sheared to prepare a PCR-free library of 150 bp. High-throughput sequencing was performed using the Illumina NovaSeq 6000 system, and a total of 2.2 GB pair-end reads was generated. Trimmonmatic v0.38 (Bolger et al. Citation2014) was employed to remove the adapters and filter low-quality reads. The chloroplast genome was assembled by using the organelle assembler NOVOPlasty v4.2.1 (Dierckxsens et al. Citation2017). The unique genes of the P. davidii chloroplast genome were annotated using CPGAVAS2 web service (Shi et al. Citation2019). The gene graphical map of the chloroplast genome was constructed using cpgview (http://www.1kmpg.cn/cpgview) (Liu et al. Citation2022). Then final chloroplast genome of P. davidii was submitted to GenBank (Accession number: OP526392).
Result
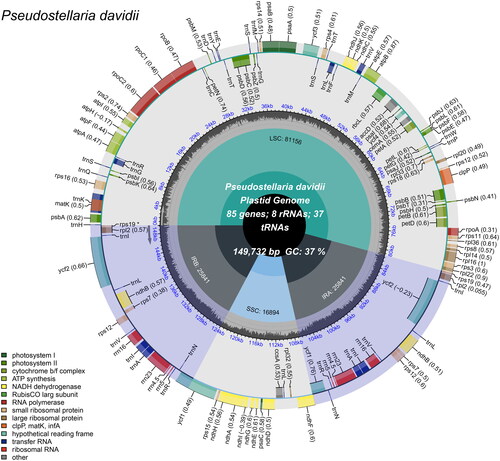
The chloroplast genome of Pseudostellaria davidii is 149,732 bp in length, with an average depth of 794.06X (Supplementary Figure 1). The genome has a conserved quadripartite structure consisting of a large single copy (LSC) region with a length of 81,156 bp, a small single copy (SSC) region of 16,894 bp, and a pair of inverted repeat regions (IRA and IRB) of 25,841 bp each. In total, 111 unique genes were predicted, including 77 protein-coding genes, 30 tRNA genes, and four rRNA genes (rrn16S; rrn23S; rrn4.5S; rrn5S) (). There were 14 genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) containing one intron and three genes (clpP, rps12 and ycf3) having two introns. Three small-exon genes (petB, petD, rps16) and one trans-spliced gene (rps12) were verified to be corrected and annotated with multiple sequence alignment (Supplementary Figure 2).
Figure 2. Schematic map of overall features of the P. davidii chloroplast genome (Genes drawn outside the outer circle are transcribed clockwise, and those inside are transcribed counter-clockwise. Genes belonging to different functional groups are color-coded. The different colored legends in the bottom left corner indicate genes with different functions. The dark grey inner circle indicates the GC content of the chloroplast genome and the presence of nodes in the LSC, SSC, IR regions).
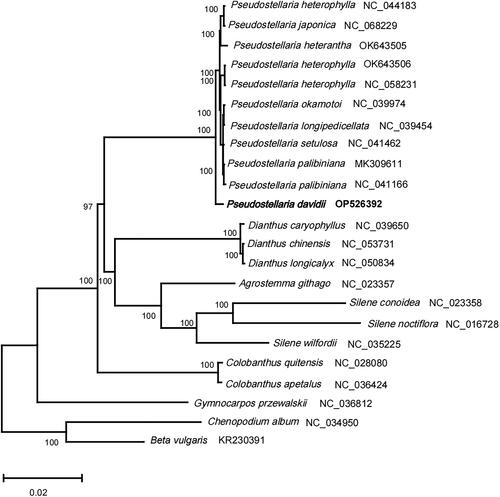
Twenty-one complete chloroplast genomes of Caryophyllaceae, and two species from Amaranthaceae (outgroup) were used for constructing Maximum Likelihood (ML) phylogenetic tree () with RAxML v8.2.12 (Stamatakis Citation2014). The alignment of 73 protein-coding genes was first created using the muscle v5 (Edgar Citation2022), and then concatenated to a super alignment with a length of 66430 bp. Species of the Pseudostellaria genus were clustered together in the phylogenetic trees with a bootstrap of 100, suggesting this genus was a confident monophyletic group. Moreover, P. davidii showed an independent sister clade to other species of the genus Pseudostellaria. This study provides the cp genome information of P. davidii, which would contribute to the species identification and phylogenetic analysis within Pseudostellaria and Caryophyllaceae species.
Figure 3. The phylogenetic position for Pseudostellaria davidii according to the ML phylogenetic tree constructed based on 23 chloroplast genomes. The following sequences were used: Pseudostellaria heterophylla NC_044183 (Kim et al. Citation2019b), Pseudostellaria japonica NC_068229, Pseudostellaria heterophylla OK_643505, Pseudostellaria heterophylla OK_643506, Pseudostellaria heterantha NC_058231, Pseudostellaria okamotoi NC_039974, Pseudostellaria longipedicellata NC_039454, Pseudostellaria setulosa NC_041462, Pseudostellaria palibiniana MK_309611 (Kim et al. Citation2019a), Pseudostellaria palibiniana NC_041166, Dianthus caryophyllus NC_039650, Dianthus chinensis NC_053731, Dianthus longicalyx NC_050834, Agrostemma githago NC_023357 and Silene conoidea NC_023358 (Sloan et al. Citation2014), Silene noctiflora NC_016728 (Sloan et al. Citation2012), Silene wilfordii NC_035225, Colobanthus quitensis NC_028080, Colobanthus apetalus NC_036424, Gymnocarpos przewalskii NC_036812, Chenopodium album NC_034950 (Hong et al. Citation2017), Beta vulgaris subsp. vulgaris KR_230391 (Stadermann et al. Citation2015). The sequences used for the tree structure are coding sequences, and the bootstrap support values are shown on the nodes.
Discussion and conclusion
In this study, the chloroplast genome sequence of Pseudostellaria davidii was assembled for the first time and the structure of this species was annotated. The phylogenetic results indicated that P. davidii showed an independent sister clade to other species of genus Pseudostellaria, and this study provided new information for the phylogenetic relationship of the Caryophyllaceae family.
Ethical approval
The material involved in the article does not involve ethical conflicts. This species is neither endangered on the CITES catalogue nor collected from a natural reserve, so it did not need specific permissions or licenses. All collection and sequencing work was strictly executed under local legislation and related laboratory regulations to protect wild resources.
Author contribution statement
Jinxin Liu and Linchun Shi conceived and designed the experiments. Hongye Zhao, Xinyi Li and Jingyi Zhao performed the experiments. Zhaolei Zhang and Yu Tian analyzed the data; Hongye Zhao and Zhaolei Zhang wrote the manuscript. Jinxin Liu revised the manuscript. All authors approved the final version and agreed to be accountable for all aspects of the work.
Supplemental Material
Download TIFF Image (64.4 MB)Supplemental Material
Download TIFF Image (23.5 MB)Supplemental Material
Download MS Word (379.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OP526392. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA903544, SRR22352177, and SAMN31807405, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Choi K, Kim J. s, Pak JH. 2001. Natural hybridization between Pseudostellaria davidii and Pseudostellaria palibiniana (Caryophyllaceae). Plant Species Biol. 16(1):39–47.
- Cui X, Hu Z, Ren S, Liu J. 2020. Pollen morphology of Chinese Pseudostellaria (Caryophyllaceae) and its systematic significance. Microsc Res Tech. 83(5):481–489.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Edgar RC. 2022. Muscle5:High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat Commun. 13(1):6968.
- Gu X, Hao D, Xiao P. 2022. Research progress of Chinese herbal medicine compounds and their bioactivities: Fruitful 2020. Chin Herb Med. 14(02):171–186.
- Hong S-Y, Cheon K-S, Yoo K-O, Lee H, Cho K-S, Suh J-T, Kim S-J, Nam J-H, Sohn H-B, Kim Y-H. 2017. Complete chloroplast genome sequences and comparative analysis of Chenopodium quinoa and C. album. Front Plant Sci. 8:1696.
- Kim Y, Heo K-I, Park J. 2019a. The second complete chloroplast genome sequence of Pseudostellaria palibiniana (Takeda) Ohwi (Caryophyllaceae): intraspecies variations based on geographical distribution. Mitochondrial DNA Part B. 4(1):1310–1311.
- Kim Y, Hong X, Jongsun P. 2019b. The complete chloroplast genome of prince ginseng, Pseudostellaria heterophylla (Miq.) Pax (Caryophyllaceae). Mitochondrial DNA B Resour. 4(2):2251–2253.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2022. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Res. 0:1–11.
- Pax F, Engler H, Prantl KJDnP 1934. Pseudostellaria. Die Naturlichen Pflanzenfamilien. ed.2.16c:318.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Sloan DB, Alverson AJ, Wu M, Palmer JD, Taylor DR. 2012. Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus silene. Genome Biol Evol. 4(3):294–306.
- Sloan DB, Triant DA, Forrester NJ, Bergner LM, Wu M, Taylor DR. 2014. A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (Caryophyllaceae). Mol Phylogenet Evol. 72:82–89.
- Stadermann KB, Weisshaar B, Holtgräwe D. 2015. SMRT sequencing only de novo assembly of the sugar beet (Beta vulgaris) chloroplast genome. BMC Bioinformatics. 16(1):295.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yang L, Feng C, Cai M, Chen J, Ding P. 2020. Complete chloroplast genome sequence of Amomum villosum and comparative analysis with other Zingiberaceae plants. Chin Herb Med. 12(4):375–383.
- Zeng X, Zhang M, Lei Y. 2016. Classification outline and geographical distribution of Pseudostellaria Pax in the world. J Plant Res Environ. 25(02):92–99.

