Abstract
In this study, the complete mitochondrial genome of Parachaeturichthys polynema was reported. The mitochondrial genome was 16,620bp in length including 13 protein-coding genes, 23 tRNAs, 2 rRNAs, and a control region. The overall contents of A, T, G, and C were 28.56%, 26.34%, 16.22%, and 28.89%, respectively. Phylogenetic analysis of 18 species in the family Gobiidae showed that P. polynema was clustered into the subfamily Gobiinae. This information will contribute to future phylogenetic studies of P. polynema and Gobiidae.
Introduction
Parachaeturichthys polynema (Bleeker,1853) is a small fish that inhabits the warm water of the Indo-West Pacific (Wu and Zhong Citation2008; Rao Citation2015). It is commonly called a tail-eyed goby with a distinctive feature with a black ocellus on the upper part of the caudal fin (). P. polynema belongs to the family Gobiidae. The Gobiidae is one of the most species-rich of fish families (Nelson et al. Citation2016), and the morphological identification and classification of gobies are still controversial (Thacker Citation2003). Molecular phylogenetics provided a novel avenue for taxonomic studies of the group (Thacker and Roje Citation2011). In this study, the complete mitochondrial genome of P. polynema is reported. We expect that the genomic data will provide an important resource for the studies on phylogenetic relationships and molecular evolution of the family Gobiidae.
Materials and methods
An adult individual of P. polynema was collected from the fisheries resource survey in the East China Sea (122°19′E, 29°0′N) in April 2020. The specimen was stored in the East China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (contact person Hanye Zhang and email [email protected]) under the voucher number ECSFRI-NMW01.
Total genomic DNA was extracted from muscle tissues using standard phenol/chloroform techniques (Sambrook et al. Citation1989). The complete P. polynema mitochondrial DNA sequence was determined with 17 pairs of primers (listed in supplementary file), which were designed based on Istigobius campbelli mitogenome sequence (GenBank accession number NC_046573), for polymerase chain reaction (PCR) amplification and sequencing. The complete mitochondrial genome sequences were assembled and annotated with the software Geneious (Drummond et al. Citation2010). We deposited the complete mitogenome sequence of P. polynema in GenBank database with accession number OK012405.
Results and discussion
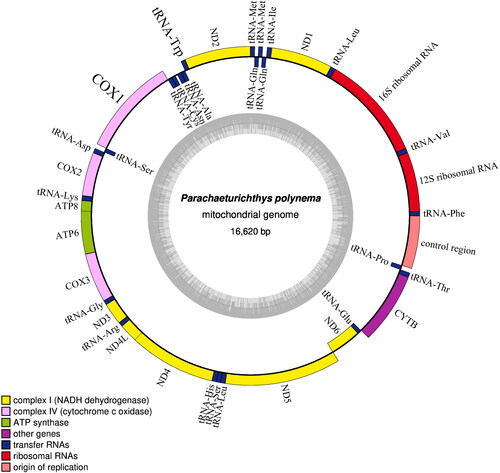
The circular mitogenome of P. polynema is 16,620 bp in length and includes 13 protein-coding genes(PCGs), 23 transfer RNA genes, 2 ribosomal RNA genes, and a control region (). The ND6 gene and nine tRNA genes are encoded on the light strand, while all of the remaining genes are located on the heavy strand. Of the 13 PCGs, 12 initiate with ATG as the start codon; the exception being the COI gene, which starts with GTG. 8 PCGs terminated the typical TAA or TAG as the stop codon, while 5 PCGs (COX2, COX3, ND3, ND4 and CYTB) ended with an incomplete T. The mitogenome contains 8 gene overlaps totalling 40 bp and ranging from 1 to 15 bp, and there were 13 intergenic regions with a total of 64 bp observed, ranging from 1 to 38 bp. P. polynema had the same characteristics as other Gobiidae species in mitochondrial genome structures, and no unique gene arrangement/gene order was found. The overall contents of A, T, G, and C were 28.56%, 26.34%, 16.22%, and 28.89%, respectively. Similar to the mitogenomes of other vertebrates, the A + T content is higher than the G + C content (Saccone et al. Citation1999; Zhu et al. Citation2013).
Figure 2. Gene map of the mitochondrial genome of P. polynema. Genes outside the circle are encoded on the heavy strand and genes inside the circle are encoded on the light stand. The inner circle indicates the GC content. Visualization was generated using the OGDRAW version 1.3.1 (Greiner et al. Citation2019).
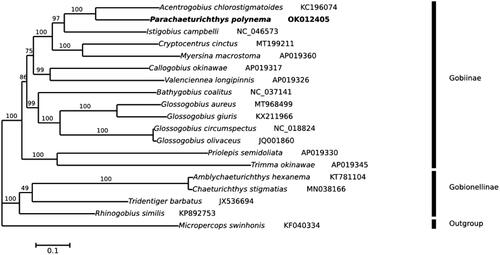
Phylogenetic analysis was performed with P. polynema and the other 17 Gobiidae species downloaded from GenBank based on 13 protein-coding genes. Using Micropercops swinhonis as an outgroup phylogenetic tree was constructed using IQ-TREE v1.6.8 (Nguyen et al. Citation2015) for maximum likelihood (ML) method with 5000 ultrafast bootstraps. In , the results suggest that P. polynema is most closely related to Acentrogobius chlorostigmatoides, and formed a monophyletic group with other Gobiinae species. Phylogenetic analysis supports Gobiinae and Gobionellinae as sister groups within the family Gobiidae, which remains controversial in the taxonomy.
Ethical approval
This study did not involve endangered or protected species. The fish samples were collected from the fisheries resource survey which was granted permission by the Ministry of Agriculture and Rural Affairs of the People’s Republic of China.
Author contributions
H. Zhang and Q. Cheng designed and conceived the study. H. Zhang collected the samples and wrote the manuscript. Q. Cheng carried out the experiments and analyzed the data. All authors revised the manuscript and agree to be responsible for all aspects of the work.
Supplemental Material
Download MS Word (16.5 KB)Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of this article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number OK012405. The associated BioProject and Bio-Sample numbers are PRJNA756399 and SAMN32983068, respectively.
Additional information
Funding
References
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S. 2010. Geneious Version 4.8.5. http://www.geneious.com/.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the world. 5th ed. New Jersey: John Wiley & Sons, Inc.; p. 331–333.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Rao Y, Veni NK, Sirisha R, Rani S. 2015. Some aspects of biology of tail eyed goby Parachaeturichthys polynema (Bleeker, 1853), off Bisakhapatnam, east coast of India. Global Journal of Biology. Agriculture Health Sci. 4(1):238–247.
- Saccone C, Giorgi CD, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 238(1):195–209.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular Cloning: A Laboratory Manual, 2nd ed. New York: Cold Spring Harbor Laboratory Press.
- Thacker CE. 2003. Molecular phylogeny of the gobioid fishes (Teleostei: Perciformes: Gobioidei). Mol Phylogenet Evol. 26(3):354–368.
- Thacker CE, Roje DM. 2011. Phylogeny of Gobiidae and identification of gobiid lineages. Syst Biodivers. 9(4):329–347.
- Wu HL, Zhong JS. 2008. Fauna Sinica, Osteichthyes, Perciformes, Gobioidei. Beijing: Science Press. p. 532–534.
- Zhu YX, Chen Y, Cheng QQ, Qiao HY, Chen WM. 2013. The complete mitochondrial genome sequence of Schizothorax macropogon (Cypriniformes: Cyprinidae). Mitochondrial DNA. 24(3):237–239.