Abstract
Phyllostachys edulis f. bicolor, a beautiful ornamental bamboo species, is a new variant of P. edulis, with yellow stems and green grooves between nodes. In this study, we assembled and annotated the complete chloroplast (cp) genome of this variety for the first time. The complete cp genome size of P. edulis f. bicolor was 139,678 bp in length and a total of 130 unique genes were annotated, including 85 protein-coding genes, 37 tRNA encoding genes, and eight rRNA encoding genes. Phylogenetic analysis results provided evidence that P. edulis f. bicolor was closely related to P. edulis ‘heterocycla’. This study contributes to better understanding of intraspecific type evolution of P. edulis.
Introduction
Phyllostachys edulis f. bicolor (Phyllostachys edulis (Carr.) H. de Lehaie f. bicolor (Nakai) G. H. Lai Citation2012) is a new variant of Phyllostachys showing color variation, and considered an excellent ornamental plant (Chen Citation2013). In contrast to the archetype, its bamboo culm is mainly yellow, but the longitudinal groove on the side of the internode branches is green, with a few thin green longitudinal stripes outside the groove. Some leaves are green with yellowish thin longitudinal stripes, and the colors of the bamboo sheath, culm sheath, and patches are lighter. The varieties are mainly distributed in south of the Huaihe River and north of Nanling (Lai Citation2012; Bu and Zhang Citation2015; Zeng et al. Citation2018). In this study, using high-throughput sequencing technology, a complete chloroplast (cp) genome of P. edulis f. bicolor was obtained for the first time to provide a reference for understanding the evolution of intraspecific types of P. edulis.
Materials
Samples were collected from an experimental bamboo forest () at Nanjing Forestry University (118.817139° E, 32.078625° N). Voucher specimens of P. edulis f. bicolor (collector: Qirong Guo, [email protected], voucher number NJFU-G2017P11) were deposited in the laboratory of the herbarium at the Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University.
Methods
Genomic DNA was extracted using a Qiagen Plant Genomic DNA Prep Kit (Sangon Biotech, Shanghai, China), sequenced on the Illumina Hiseq 2500 platform, and deposited to NCBI under the record number SRR18455992. Data quality control was performed using fastp (Chen et al. Citation2018), and Novoplasty (v4.0) was used to assemble high-quality paired data into a complete cp genome. Genome annotation was performed using GeSeq (Tillich et al. Citation2017). Alignment was performed using the P. edulis cp genome sequence combined with manual manipulation, using Geneious R8 (Biomatters Ltd, Auckland, New Zealand) for correction. tRNA genes were further validated using the tRNAscan-SE online web server (Schattner et al. Citation2005). A gene map of the annotated P. edulis f. bicolor cp genome was drawn online using CPGview (Liu et al. Citation2023), and the annotated P. edulis f. bicolor cp genome sequence was submitted to NCBI (accession number: OM084949). To confirm the phylogenetic position of P. edulis f. bicolor, we obtained 11 published Phyllostachys varieties cp genomes from NCBI, and four Phyllostachys varieties cp genomes from the China National GeneBank DataBase (CNGBdb). The phylogeny was inferred using the maximum-likelihood (ML) method. MAFFT software was used for sequence alignment (Katoh and Standley Citation2013), and the phylogenetic tree was constructed using IQ-TREE (Minh et al. Citation2020). The best-fitting model was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017) and implemented in IQ-TREE.
Results
Analysis of chloroplast genome characteristics
The full length of the P. edulis f. bicolor cp genome () of 139,678 bp is comprised of a large single-copy region (LSC, 83,212 bp), small single-copy region (SSC with 12,870 bp), and two inverted repeat regions (IR with 21,798 bp). The GC content of the total genome was 38.9%, and the GC content of the LSC, SSC, and IR regions was 37.0%, 33.2%, and 44.2%, respectively. There were 130 unique genes, including 85 protein-coding genes, 37 tRNA encoding genes, and eight rRNA encoding genes. According to their function, all annotated genes were divided into four main categories: genes related to photosynthesis (n = 45), genes related to self-replication (n = 75), genes encoding other proteins (n = 5), and genes of unknown function (n = 5), with 19 of them (ndhB, rps7, rps15, rps19, rpl2, rpl23, ycf2, trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC, rrn4.5, rrn5, rrn16, and rrn23) being double-copy genes.
Figure 2. Schematic map of overall features of the chloroplast genome of Phyllostachys edulis f. bicolor.
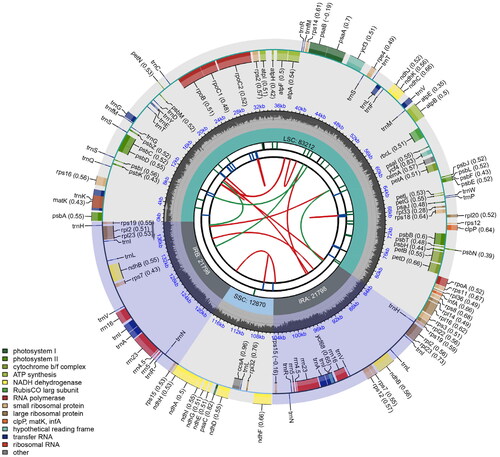
The map contains six circles. From the center going outward, the first circle shows the forward and reverse repeats connected with red and green arcs, respectively. The second circle and the third circle show tandem repeats and microsatellite sequences marked with short strips, respectively. The fourth circle indicates the position of LSC, SSC, IRA, and IRB, and the fifth circle indicates the GC content. The genes outside the sixth circle are transcribed counterclockwise, while the genes inside are transcribed clockwise. The genes were colored based on their functional categories.
Phylogenetic analysis of Phyllostachys edulis f. bicolor
The phylogenetic relationships of some P. edulis varieties are described using phylogenetic analysis. P. edulis ‘heterocycla’, P. edulis f. bicolor, P. edulis ‘pachyloen’, P. edulis curviculmis, and P. angusta formed a small branch, with P. edulis. P. edulis and P. angusta each gathered into one branch, P. edulis f. bicolor and P. edulis ‘heterocycla’ clustered together, and P. edulis curviculmis and P. angusta forming another cluster. This indicated that they were closely related to each other and had notable collinearity in the evolution of intraspecific types of Phyllostachys ().
Figure 3. Phylogenetic relationships between Phyllostachys edulis f. bicolor and some other species of Phyllostachys Varieties. Bootstrap support values are given at the nodes.
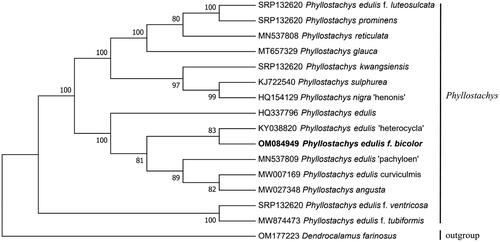
The phylogenetic tree is constructed by the ML method. The sequences used for tree construction are as follows: P. edulis f. luteosulcata (SRP132620). P. prominens (SRP132620). P. kwangsiensis (SRP132620). P. edulis f. ventricose (SRP132620). P. reticulata (MN537808; Huang et al. Citation2019). P. glauca (MT657329; Cao et al. Citation2020). P. sulphurea (KJ722540; Gao and Gao Citation2016). P. nigra ‘henonis’ (HQ154129). P. edulis (HQ337796). P. edulis ‘heterocycla’ (KY038820; Dan et al. Citation2017). P. edulis f. bicolor (OM084949). P. edulis ‘pachyloen’ (MN537809; Huang et al. Citation2019). P. edulis curviculmis (MW007169; Gao et al. Citation2021). P. angusta (MW027348; Yu et al. Citation2021). P. edulis f. tubiformis (MW874473; Liu et al. Citation2022). Dendrocalamus farinosus (OM177223; Pei et al. Citation2022).
Discussion and conclusions
China’s Phyllostachys resources are rich; there are several varieties with significant phenotypic genetic difference among them (Zhang et al. Citation2007). In this study, we successfully assembled the complete cp genome of Phyllostachys edulis f. bicolor for the first time and selected the complete cp genome data of 15 other Phyllostachys varieties for phylogenetic analysis. The results showed that P. edulis f. bicolor was closely related to P. edulis ‘heterocycla’. The findings were consistent with those of previous studies on P. edulis curviculmis (Gao et al. Citation2021), where P. edulis f. bicolor formed a small branch with P. edulis curviculmis and P. edulis ‘pachyloen’, all of which are members of the Phyllostachys genus. Moreover, the cp genome size and structure of P. edulis f. bicolor and P. edulis curviculmis were the same, which further indicated the genetic relationship between the two. Elucidation of the complete cp genome of P. edulis f. bicolor provides important data for the phylogeny of Phyllostachys varieties that will help us in better understanding of the evolution of intraspecific types of Phyllostachys.
Author contributions
Qirong Guo contributed to the study concept and design; Bo Hu and Rou Wan collected the samples and performed the experiments; Wenxuan Jing analyzed and interpreted the data; all authors read and approved the final manuscript; and that all authors agree to be accountable for all aspects of the work.
Ethics statement
We confirmed that all the research meets ethical guidelines and adheres to the legal requirements of the study country. This study did not involve any ethical issues, so no ethics committee or relevant permissions were required.
Supplemental Material
Download MS Word (285.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, under the accession number OM084949. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA818118, SRR18455992, and SAMN26816507, respectively.
Additional information
Funding
References
- Bu LQ, Zhang XM. 2015. Ornamental features of Phyllostachys spp. and its landscaping design. World Bamboo Rattan. 13(4):33–36.
- Cao B, Ge TT, Ding SX, Guo CC. 2020. The complete chloroplast genome of Phyllostachys glauca (Bambusoideae), a dominant bamboo species in limestone mountains endemic to China. Mitochondrial DNA B Resour. 5(3):3193–3194.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Chen TG. 2013. Phyllostachys edulis f. bicolor. World Bamboo Rattan. 11(2):12–13.
- Dan Z, Yuan L, Li ZG. 2017. The complete chloroplast genome sequence of Phyllostachys heterocycla, a fast-growing non-timber bamboo (Poaceae: Bambusoideae). Conserv Genet Resour. 9(2):217–219.
- Gao J, Gao LZ. 2016. The complete chloroplast genome sequence of the Phyllostachys sulphurea (Poaceae: Bambusoideae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):983–985.
- Gao LQ, Li YL, Zhang WG, Yang GY. 2021. The complete chloroplast genome of Phyllostachys edulis curviculmis (Bambusoideae): a newly ornamental bamboo endemic to China. Mitochondrial DNA B Resour. 6(3):941–942.
- Huang NJ, Li JP, Yang GY, Yu F. 2019. Two plastomes of Phyllostachys and reconstruction of phylogenic relationship amongst selected Phyllostachys species using genome skimming. Mitochondrial DNA B Resour. 5(1):69–70.
- Kalyaanamoorthy S, Minh B, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lai GH. 2012. A reconsideration of taxonomic position of some infraspecific mutants in the genus Phyllostachys (Bambusoideae). J Anhui Agric Sci. 40(8):4621–4625.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGview: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Liu XM, Liu L, Li LB, Yue JJ. 2022. The complete chloroplast genome of Phyllostachys edulis f. tubiformis (Bambusoideae): a highly appreciated type of ornamental bamboo in China. Mitochondrial DNA B Resour. 7(1):185–187.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Pei JL, Wang Y, Zhuo J, Gao HB, Vasupalli N, Hou D, Lin XC. 2022. Complete chloroplast genome features of Dendrocalamus farinosus and its comparison and evolutionary analysis with other Bambusoideae species. Genes. 13(9):1519–1533.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server):W686–W689.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yu Z, Lin YZ, Tao W, Guo FC, Yi FH, Jin JY. 2021. The complete chloroplast genome of Phyllostachys angusta McClure (Poaceae). Mitochondrial DNA B Resour. 6(1):187–188.
- Zeng QN, Yu L, Cheng P, Wang HX, Peng JS. 2018. A study of culm color characteristic and type classification of Phyllostachys edulis cv. Tao Kiang. World Bamboo Rattan. 16(4):32–35.
- Zhang SF, Ma QX, Ding YL. 2007. Genetic diversity analysis of moso bamboo based on morphological characterization. J Bamboo Res. 7(3):16–21.

