Abstract
Tamarix L. is of great ecological and economic significance in arid desert ecosystems. This study reports the complete chloroplast (cp) genomic sequences of T. arceuthoides Bunge and T. ramosissima Ledeb., which are currently unknown, by high-throughput sequencing. The cp genomes of T. arceuthoides 1852 and T. ramosissima 1829 were 156,198 and 156,172 bp in length, respectively, and contained a small single-copy region (SSC: 18,247 bp), a large single-copy region (LSC: 84,795 and 84,890 bp, respectively), and a pair of inverted repeat regions (IRs: 26,565 and 26,470 bp, respectively). The two cp genomes possessed 123 genes arranged in the same order, including 79 protein-coding, 36 tRNA, and eight rRNA genes. Of these, 11 protein-coding genes and seven tRNA genes contained at least one intron. The present study found that Tamarix and Myricaria are sister groups with the closest genetic relationship. The obtained knowledge could provide useful information for future phylogenetic, taxonomic, and evolutionary studies on Tamaricaceae.
Introduction
Tamarix L. contains approximately 100 species that are primarily distributed in the arid and semi-arid areas of continental Asia and North Africa, along with intermittent distribution along the west coast of South Africa and parts of Europe (Zhang and Zhang Citation1990). There are 20 Tamarix species found in China, of which 16 species are known to propagate in Xinjiang (Liu Citation2014). Certain Tamarix species have been employed in ecological restoration projects to achieve objectives, including wind prevention, sand fixation, soil and water conservation, and climate regulation, showing great ecological and economic value in the maintenance of arid desert ecosystems (Baum Citation1978; Liu Citation2014). However, due to their similar morphological characteristics, the identification among this genus is frequently mistaken, which affects the effective development and utilization of some species in the genus Tamarix.
The complete chloroplast (cp) genomes present an effective means of improving the rate of species identification and has been developed as a tool for plant phylogenetic studies at different taxonomic levels (Zhou et al. Citation2021; Wang et al. Citation2022). Till date, three Tamarix cp genomes, including those from two species and one unverified sequence (MN726883) have been published (Pang et al. Citation2021; Wang et al. Citation2021). This paucity of data has severely limited relevant research on this genus Tamarix. Here, we sequenced the cp genomes of T. arceuthoides Bunge 1852 and T. ramosissima Ledeb. 1829 (), to conduct comprehensive research on the Tamarix cp genome, and serve as a reference for subsequent phylogenomic studies of the Tamarix.
Figure 1. The individual, raceme, flower and capsule photograph of T. arceuthoides (B, D, F, and H) and T. ramosissima (A, C, E, and G), photo by Xiyong Wang at Turpan Eremophytes Botanical Garden. Main identifying traits: When T. ramosissima flowers, the petals are completely open, and the petals do not persist at fruit stage, but fall off directly and T. arceuthoides flowers cup shaped when flowering, petals persistent in fruit, attached to the base of the fruit.

Materials
Samples of T. arceuthoides and T. ramosissima were collected from the Turpan Eremophytes Botanical Garden, Chinese Academy of Sciences, Turpan, Xinjiang, China (latitude 89°19′27.78″, longitude 42°85′71.92″ and latitude 89°11′22.56″, longitude 42°51′12.96″, respectively), and voucher specimens were deposited at the Specimen Museum of Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences (XJBI, Li Wenjun, [email protected]) with collection number LWJ-W-4, LWJ-W-7 and identifier Xi. Y. Wang. All materials utilized in the present study are listed in Supplementary Table.
Methods
Leaf samples were dried in silica gel and stored at −20 °C for DNA extraction, which was performed using a plant genome extraction kit (DP320) obtained from Tiangen Biochemical Technology (Beijing, China) as per the manufacturer’s instructions. Extracted DNA was sequenced with 2 × 150 bp paired-end reads on the Illumina HiSeq X Ten platform at the Molecular Biology Experiment Center, Germplasm Bank of Wild Species in Southwest China.
Paired-end reads were assembled using GetOrganelle v. 1.7.1 (Jin et al. Citation2020). A complete circular assembly graph was checked and further extracted using Bandage version 0.8.1 (Wick et al. Citation2015). The genomes were automatically annotated using CpGAVAS (Liu et al. Citation2012) and PGA (https://github.com/quxiaojian/PGA), prior to manual adjustments using Geneious version 9.1.7 (Kearse et al. Citation2012), with the cp genome of T. taklamakanensis (MW125612) as a reference. The sequence data of T. arceuthoides and T. ramosissima are publicly available in the GenBank (https://www.ncbi.nlm.nih.gov/) under accession numbers ON620259 and ON620260. Organellar Genome Draw (OGDRAW) (Lohse et al. Citation2007) was used to illustrate the circular genome map.
To further explore the phylogenetic relationship of among Tamarix and its neighboring genus, maximum-likelihood (ML) analyses were conducted using RAxML-HPC v. 8 (Stamatakis Citation2014) with 1000 bootstrap replicates based on 11 cp genomes (after removing one inverted repeat (IR) region), including six Tamarix sequences, four Myricaria sequences, and Reaumuria songarica as outgroup (Supplementary Table). The evaluation of the most appropriate substitution models and the construction of the phylogenetic tree were both carried out on the CIPRES Science Gateway portal (Miller et al. Citation2011).
Results
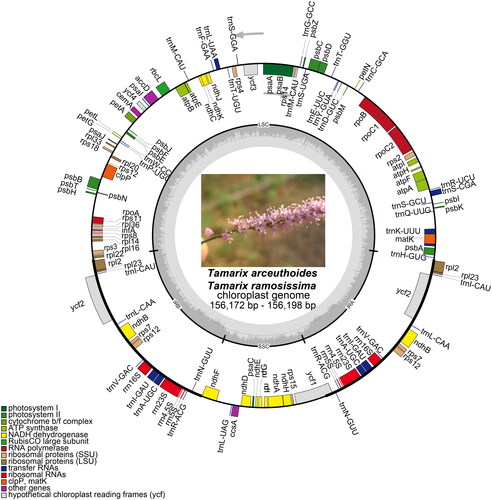
The new cp genomic sequences were found to occupy a circular confirmation, 156,172 and 156,198 bp in length, respectively, comprising a small single-copy region (SSC: 18,247 bp) as well as a large single-copy region (LSC: 84,795 and 84,890 bp, respectively) that were separated by a pair of inverted repeat regions (IRs: 26,565 and 26,470 bp, respectively). Both the cp genomes possessed 123 genes that were arranged in the same order, including 79 protein-coding genes, 36 tRNA genes, and eight rRNA genes. Of these, 11 protein-coding and seven tRNA genes contained at least one intron (). These two sequences had 36.4% and 36.5% GC content, respectively. Comparison of the two cp genomes to previously published data revealed a high level of gene synteny with one publicly available genome data sets from T. taklamakanensis (MW125612).
Figure 2. Chloroplast genome maps for T. arceuthoides and T. ramosissima. Genes on the inside of the circle are transcribed clockwise and those on the outside are transcribed counterclockwise. The darker gray inner circle corresponds to the GC content, whereas the lighter gray indicates the AT content. Different colors represent different functional genes. The thick lines of the large circle indicate the extent of the inverted repeat regions (IRa and IRb) that separate the genome into small single-copy (SSC) and large single-copy (LSC) regions, respectively.
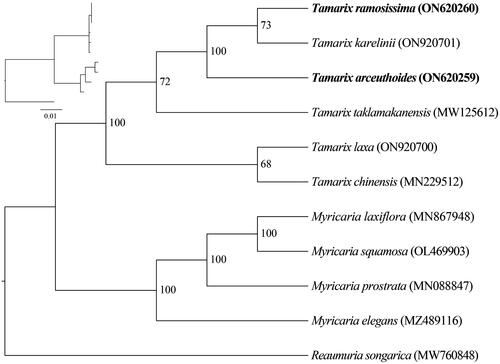
The phylogenetic tree analysis revealed that Tamarix and Myricaria were sister groups (BS = 100%). Tamarix was divided into three main lineages, the new sequences T. arceuthoides and T. ramosissima clustered with T. karelinii together ().
Figure 3. Phylogenetic trees inferred from maximum-likelihood (ML) analyses based on 12 complete chloroplast genomes using Reaumuria songarica as an outgroup with 1000 bootstraps replicates. The numbers above the branches indicate the bootstrap values. The following sequences were used: Reaumuria songarica MW760848 (Duan Y et al. 2021), Myricaria elegans MZ489116 (Su T and Han M 2021), Myricaria laxiflora MN867948 (Wang et al. Citation2020), Myricaria prostrata MN088847 (Chi X 2020), Myricaria squamosa OL469903 (Yu L 2022), Tamarix chinensis MN229512 (Chi Citation2019), Tamarix karelinii ON920701 (Song S 2022), Tamarix laxa ON920700 (Song S 2022), and Tamarix taklamakanensis MW125612 (Yang T 2020).
Discussion and conclusions
We report the cp genomes of T. arceuthoides and T. ramosissima. The structures obtained for the two cp genomes in this study are consistent with previous findings (Pang Citation2021). Our study demonstrates that plastome studies can provide useful information for future phylogenetic, taxonomic, and evolutionary studies on Tamaricaceae. However, the complete cp genomes are not distinct between the T. arceuthoides and T. ramosissima, indicating that the molecular identification of the genus Tamarix might require to select highly variable regions in cp genomes, or add additional nrDNA sequences.
Author contributions
W Yan and XY Wang conceived and designed the study. XY Wang provided materials for this study. QM Cao and XY Wang analyzed the sequence data and wrote the manuscript. W Yan revised the manuscript. All authors approved the final manuscript.
Ethical approval
The materials used in this study were not related to protected species and were within the limits of the relevant national laws. We obtained permission from the Turpan Desert Botanical Garden, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences to collect plant specimens.
Supplemental Material
Download JPEG Image (256.8 KB)Supplemental Material
Download JPEG Image (170.7 KB)Supplemental Material
Download JPEG Image (256.9 KB)Supplemental Material
Download PNG Image (22.4 KB)Supplemental Material
Download PNG Image (22.5 KB)Supplemental Material
Download MS Word (16.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under accession nos. ON620259 and ON620260. The associated BioProjects are PRJNA890384 and PRJNA890011; SRA are SRR21901739 and SRR21889044; and the Bio-Sample numbers are SAMN31264515 and SAMN31266535, respectively.
Additional information
Funding
References
- Baum BR. 1978. The genus Tamarix. Jerusalem: The Israel Academy of Sciences and Humanities.
- Chi XF, Zhang FQ, Chen SL. 2019. The complete chloroplast genome of Myricaria prostrata, a threatened plant in the Qinghai–Tibetan Plateau. Mitochondrial DNA Part B. 4(2):2637–2638.
- Jin JJ, Yu WB, Yang JB, Song Y, Claude W. dePamphilis, Yi T S, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome biology. 21(1):1–31.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu MT. 2014. The collection introduces the species genus Tamarix and its supporting index in China. Urumqi: Xinjiang People’s Publishing House.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- Miller MA, Pfeiffer W, Schwartz T. 2011. The CIPRES science gateway: a community resource for phylogenetic analyses. Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery; p. 1–8.
- Pang X-A, Jiao P-P, Yang T-G, Liu H. 2021. Complete chloroplast genome sequence of Tamarix taklamakanensis (Tamaricaceae). Mitochondrial DNA B Resour. 6(11):3295–3296.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang N, Chen S, Xie L, Wang L, Feng Y, Lv T, Fang Y, Ding H. 2022. The complete chloroplast genomes of three Hamamelidaceae species: comparative and phylogenetic analyses. Ecol Evol. 12(2):e8637.
- Wang L, Wang L, Guo ZH. 2021. The complete chloroplast genome of Tamarix ramosissima and comparative analysis of Tamaricaceae species. Biol Plant. 65:237–245.
- Wang Q, Zhang SD, Ding B, Zhu X, Deng HP. 2020. The complete chloroplast genome of Myricaria laxiflora (Tamaricaceae): an endemic and endangered species from China. Mitochondrial DNA Part B. 5(2):1153–1154.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Zhang PY, Zhang YJ. 1990. Flora of China. Vol. 50. Beijing: Science Press; p. 142–177.
- Zhou M, Yan M, Ying Z, He X, Ge Y, Cheng R. 2021. Characterization of the complete chloroplast genome of Oxalis corymbosa DC. (Oxalidaceae), a medicinal plant from Zhejiang Province. Mitochondrial DNA B Resour. 6(3):1138–1140.
