Abstract
Abies ernestii var. salouenensis (Bordères & Gaussen) W. C. Cheng & L. K. Fu is endemic to southwest China, including the southeastern Tibetan Plateau and the northwestern Yunnan Province. The taxonomic relationships between A. ernestii var. salouenensis and two other closely related fir species (A. chensiensis Tiegh. and A. ernestii Rehd.) still need to be determined. Here, we report for the first time the whole chloroplast genome of A. ernestii var. salouenensis. Its genome is 121,759 bp long and is characterized by a circular structure with 68 peptide-encoding genes, 16 tRNAs, six ORFs, and four rRNAs. We also identified 70 microsatellite repeat sequences and 14 tandem repeat sequences in the chloroplast genome of A. ernestii var. salouenensis. Comparative genome analysis indicated considerable variation in ycf1 and ycf2. Phylogenetic analysis supported the monophyly of A. ernestii var. salouenensis, A. chensiensis Tiegh., and A. ernestii Rehd. The relationships among them should be surveyed using more samples at the species level. This study will facilitate taxonomic studies and the development of suitable chloroplast markers for fir species.
Introduction
Abies ernestii var. salouenensis (Bordères & Gaussen) W. C. Cheng and L. K. Fu is endemic to southwest China, including the northwest Yunnan Province and the southeastern Tibetan Plateau () (Kuan Citation1981; Farjon Citation1990). It serves as an essential habitat for many plants and animals, making it an ecologically significant component of the cold-temperate woods (Farjon and Rushforth Citation1989). To date, the complete chloroplast genome features of Abies ernestii var. salouenensis has never been investigated.
Figure 1. Abies ernestii var. salouenensis. (A) Plant; (B) mature cones; (C) pollen cones; (D) leaves; (E) vegetative buds. M. These images are from Christian (Citation2021) and photography by Yi-Zhen Shao.

Species delimitation of A. ernestii var. salouenensis and two other closely related species (A. chensiensis Tiegh. and A. ernestii Rehd.) has been a contentious issue for a long time. (Suyama et al. Citation2000; Xiang et al. Citation2009; Semerikova et al. Citation2011, Citation2018; Shao and Xiang Citation2015). Unlike other fir species, these three closely related fir species have particular habitat feature such as lower altitude ranges, relatively arid habitat, and neutral to slightly alkaline soils (Liu Citation1971; Farjon Citation2001). A series of studies using morphological characteristics and molecular markers have been conducted to address this taxonomic problem (Xiang et al. Citation2004, Citation2015). Unfortunately, the three closely related fir species have never been simultaneously surveyed using the recommended high-resolution markers (e.g. chloroplast genomes) (Liepelt et al. Citation2010; Aguirre-Planter et al. Citation2012; Xiang et al. Citation2009, Citation2018). Consequently, several distinct taxonomic pairings have been proposed: Abies ernestii var. salouenensis was once thought to be a subspecies or variety of A. chensiensis (Rushforth Citation1984; Shao and Xiang Citation2015); and A. ernestii var. salouenensis, A. ernestii, and A. chensiensis were treated identically by Handel-Mazzetti (Citation1929) and Dallimore and Jackson (Citation1966). Recently, complete chloroplast genome data have emerged as one of the most effective indicators for distinguishing taxonomically complex groups (Shao et al. Citation2022). Thus, a more thorough approach utilizing chloroplast genomes is required to elucidate the relationships among these three closely related fir species.
In this study, we constructed and sequenced the chloroplast genome of A. ernestii var. salouenensis, and then conducted a comparative analysis with A. chensiensis and A. ernestii. This will facilitate taxonomic studies and the development of suitable chloroplast markers for fir species.
Materials and methods
Plant collection and DNA extraction
The leaves of A. ernestii var. salouenensis were collected by Qiao-Ping Xiang from Deqen County, Yunnan Province, China (E98.90°, N28.18°). A voucher specimen (Voucher Number: BMXDC) was preserved in the herbarium of the Institute of Botany, CAS (PE) (http://pe.ibcas.ac.cn, Qin Ban, [email protected]). The entire genome of A. ernestii var. salouenensis was extracted using an EZNA Plant DNA Extraction Kit (OMEGA, USA).
Genome sequencing, assembly, and annotation
A genomic library was constructed using the TruSeq Nano DNA Sample Prep Kit (Illumina, USA) according to the manufacturer’s protocol. The libraries were 150 bp long and sequenced on an Illumina HiSeq X platform. GeSeq, tRNAscan-SE v1.3.1, and a CLC de novo assembler (CLC Bio, Aarhus, Denmark) were used for further alignment, assemble, and annotation of reads (Schattner et al. Citation2005; Tillich et al. Citation2017). The readings were edited using quality restriction of Q5 and N > 10% to ensure excellent quality. These reads were alligned to the reference sequence using Velvet (Zerbino and Birney Citation2008). To match the gene predictions, we checked all the start/stop codons and intron/exon boundaries in Sequin and Geneious (Kearse et al. Citation2012; Lohse et al. Citation2013). Finally, the sequences were annotated by comparing them with published genomes. The overall coverage depth of the chloroplast genome assembly of Abies ernestii var. salouenensis was 124× (Figure S1). The associated GenBank accession number was MH706708.
Repeat sequences detection and comparative genomic analysis
The MISA program was used to examine simple sequence repeats (SSRs). The REPuter website was used to survey long repeats (Kurtz et al. Citation2001). The maximum and minimum computed repeats were 50 bp and 30 bp, respectively. The Hamming distance was set to three. Next, we examined the complete chloroplast genomes of A. ernestii var. salouenensis (MH706708), A. ernestii (MH706707), and A. chensiensis (MH047653 and MH706706), which was available in the NCBI Database. mVista was used for comparative genomic analysis in the Shuffle-LAGAN mode (Frazeret al. Citation2004). Ten repeating units for mono-nucleotides, five for di-nucleotides, four for tri-nucleotides, three for tetra-nucleotides, three for penta-nucleotides, and three for hexa-nucleotides were set as the appropriate repeat units.
Phylogenetic analysis
Phylogenetic analysis was performed using 23 reported chloroplast genomes of fir species and Keteleeria davidiana (Bertr.) Beissn. as an outgroup. These 23 genomes represented the main clades of the genus Abies, and all are closely related species of A. ernestii var. salouenensis (Xiang et al. Citation2018). Whole genomes were aligned using MAFFT v.7 (Katoh and Standley Citation2013). We performed and visualized the maximum likelihood (ML) analysis (bootstrap search steps = 1000) using RAxML v.8.1 and FigTree v.1.4 (Bootstrap search steps = 1000), respectively (Stamatakis Citation2014). The details of the 23 complete chloroplast genomes are listed in Table S1.
Results and discussion
Chloroplast genome features of A. ernestii var. salouenensis
The whole chloroplast genome of A. ernestii var. salouenensis displayed a circular DNA molecule of 121,759 bp in length. The genome displayed a significant A/T bias of 61.70%, similar to that of other fir species (Shao et al. Citation2022). It possessed a normal quadripartite structure with two IR regions (264 bp), one long single-copy region (LSC) region (67,155 bp), and one short single-copy region (SSC) region (54,076 bp). We identified 3 open reading frames (ORFs), 53 protein-coding genes, and 16 tRNA genes in the LSC. The SSC region was home to all four rRNAs. The IR region was 264 bp long and contained the trnI-CAU and trnT-GGU genes. We identified 113 genes, including 68 peptide-encoding genes (CDS), 16 transfer RNAs (tRNA), 6 ORFs, and 4 ribosomal RNAs (rRNA) (Table S2, ). The clpP and ycf3 contained two introns, whereas only one intron occurred in atpF/rps12/trnL-UAA/rpl16/petB/trnV-UAC/petD/trnA-UGC/rpoC1/trnG-GCC/trnK-UUU (, Figure S2). Similar to other fir species, there was a palindromic inverted 1180 bp-repeat (ycf12/trnS/psaM/trnG) in the 52-kb inversion points, and the ndh genes were absent (Shao et al. Citation2022). The widespread absence of ndh genes in the Pinaceae can be explained by their high substitutability (Blazier et al. Citation2011).
Figure 2. Schematic map of overall features of the chloroplast genome of Abies ernestii var. salouenensis. The circular map of the chloroplast genome was generated using CPGview (Liu et al. Citation2023). Genes shown outside the circle are transcribed clockwise, and genes inside are transcribed counter-clockwise. Genes belonging to different functional groups are color-coded. The darker gray in the inner corresponds to the GC content, and the lighter gray to the AT content.
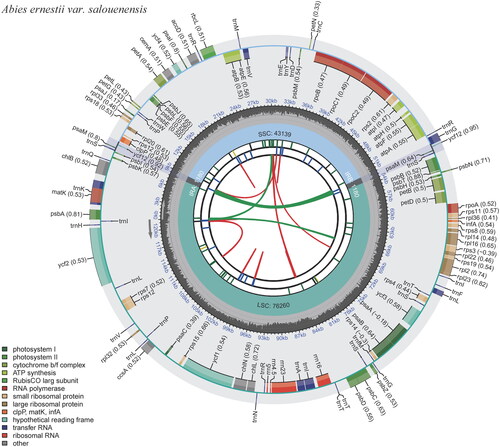
Table 1. List of genes encoded in Abies ernestii var. salouenensis chloroplast genomes.
Repeat sequence analysis
Due to their excellent reliability, simple sequence repeats (SSRs) have been utilized extensively in phylogenetic research (Kaur et al. Citation2015). We identified 70 SSRs containing mononucleotide repeats (44), dinucleotide repeats (14), trinucleotide repeats (2), tetranucleotide repeats (8), and pentanucleotides repeats (2) in the chloroplast genome of A. ernestii var. salouenensis (Figure S3). Mononucleotide repeats (62.86%) were most abundant, followed by pentanucleotides (2.86%) and trinucleotide repeats (2.86%). This indicates that mononucleotide repeats contributed the most to genetic diversity. We identified two kinds of trinucleotide SSRs (ATT/AAT), twelve kinds of tetranucleotide SSRs (AAAT/ATCT/AAAG/AGAT/ATTT/ACCT/AACC/GGTT/ATCC/CTTT/ATGG/AGGT), and four kinds of pentanucleotide SSRs (ATTCG/ATGTT/AACAT/AATCG/) (Figure S3). In addition, 51 long repeats, including palindromic repeats (15), tandem repeats (14), and forward repeats (22), were identified (Figure S3).
Comparative genome analysis
The IR region was significantly more conserved than the LSC and SSC sections in the comparative genome analysis. A significant bulk of the genetic diversity in these three fir species was contained in the noncoding and intergenic regions. This may explain the absence of DNA markers in closely related fir species in published research (Shao and Xiang Citation2015; Xiang et al. Citation2018; Shao et al. Citation2020). In addition, we compared the genomes of these three species to aid the search for applicable DNA barcodes (Figure S4). In our analysis, only ycf1 and ycf2 were characterized by considerable variation and could be suggested as potential chloroplast markers (Figure S4). As reported by Dong et al. (Citation2015), ycf1 is rich in short repeats and may be the most effective plastid barcode. In addition, the widely used markers in fir species (e.g. rpl16, matK, trnC-D, rps18, trnS-G) were lack of resolution among these three fir species (Shao et al. Citation2018).
Phylogenomic analysis
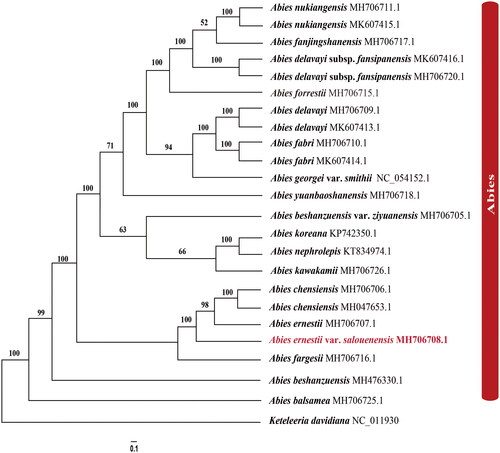
The rapid development of new approaches has significantly expanded the available chloroplast genome data. To infer the phylogenetic relationships between A. ernestii var. salouenensis, A. ernestii, and A. chensiensis, we selected 23 reported chloroplast genomes of fir species, using Keteleeria davidiana as an outgroup (). Phylogenetic analyses showed that Abies species formed a monophyletic lineage (BSML = 100). Within the genus Abies, the species from North America (A. balsamea (L.) Mill.) and East Asia formed a clear sister lineage (BSML = 100). In the East Asian clade, Abies ernestii var. salouenensis, A. ernesti, and A. chensiensis formed a monophyletic lineage (BSML = 100). Our results further indicated that A. ernesti was much more similar to A. chensiensis rather than its subspecies, Abies ernestii var. salouenensis (BSML = 96) (). Therefore, the status of A. ernestii var. salouenensis as a variety of A. ernestii might not be supported. However, the phylogenetic relationships among the three species were polyphyletic (). This could be explained by the limited sample size (N = 1 or 2) and the influence of hybridization (Xiang et al. Citation2015; Shao et al. Citation2020; Shao et al. Citation2022). Considering the above results, our phylogenomic analyses proved that chloroplast genome data are an effective indicator for distinguishing closely related fir species. Future studies should include additional individuals at the species level.
Figure 3. The best Maximum likelihood (ML) phylogram inferred from 23 chloroplast genomes in Abies, with Keteleeria davidiana as an outgroup (bootstrap values are indicated on the branches). The following sequences were used: A. nukiangensis Cheng et L. K. Fu MH706711 and MK607415 (Shao et al. Citation2020), A. fanjingshanensis W. L. Huang, Y. L. Tu and S. Z. Fang MH706717 (Guo et al. Citation2019), A. delavayi subsp. fansipanensis (Q.P.Xiang, L.K.Fu and Nan Li) Rushforth MH706720 and MK607416 (Shao et al. Citation2020), A. forrestii C. C. Rogers MH706715 (Dong et al. Citation2021), A. delavayi Franch. MH706709 and MK607413 (Shao et al. Citation2020), A. fabri (Mast.) Craib MH706710 and MK607414 (Shao et al. Citation2020), A. georgei var. smithii (Viguie et Gaussen) Cheng et L NC_054152 (Li et al. Citation2021), A. yuanbaoshanensis Y. J. Lu & L. K. Fu MH706718 (Zhang et al. Citation2020), A. beshanzuensis var. ziyuanensis (L. K. Fu & S. L. Mo) L. K. Fu & Nan Li MH706705 (Fu et al. Citation2019), A. koreana E. H. Wilson KP742350 (Yi et al. Citation2015), A. nephrolepis (Trautv.) Maxim. KT834974 (Yi et al. Citation2016), A. kawakamii (Hayata) T. Ito MH706726 (Shao et al. Citation2019), A. chensiensis MH706706 and MH047653 (Liu et al. Citation2018; Su et al. Citation2019), A. ernestii MH706707 (Shao et al. Citation2022), A. fargesii Franch. MH706716 (Guo et al. Citation2019), A. beshanzuensis M. H. Wu MH 476330 (Shao et al. Citation2018), A. balsamea (L.) Mill. MH 706725 (Wu et al. Citation2019), and Keteleeria davidiana NC_011930 (www.ncbi.nlm.nih.gov/).
This study offers new evidence and validates the potential reliability of using complete chloroplast genomes for problematic firs. These findings indicate a vital genetic resource for ecologically significant fir species.
Ethics statement
All plant-related procedures in this study adhered to the Plant Use guidelines of Henan Agriculture University and the Institute of Botany, Chinese Academy of Sciences.
Author contributions
Yi-Zhen Shao, Peng-Fei Zhao, and Si Wu contributed to the conception and design of the study. Yi-Zhen Shao, Zhao Wang, and Wen-Jun Liu contributed to the analysis and interpretation of the data. Yi-Zhen Shao, Zhao Wang, and Wen-Jun Liu wrote the first version of the manuscript. Peng-Fei Zhao and Si Wu critically reviewed and modified the article regarding its intellectual content. All authors read, discussed, and approved the final version, and all agree to be accountable for all aspects of the work.
Supplemental Material
Download TIFF Image (6.6 MB)Supplemental Material
Download TIFF Image (5.5 MB)Supplemental Material
Download TIFF Image (6 MB)Supplemental Material
Download TIFF Image (15.9 MB)Supplemental Material
Download MS Word (4.2 MB)Acknowledgements
We appreciate the assistance of Professor Qiao-Ping Xiang from the Chinese Academy of Sciences’ Institute of Botany with the fieldwork.
Disclosure statement
There are no conflicts of interest disclosed by the authors. Authors are solely responsible for the accuracy and integrity of the information.
Data availability statement
The genomic sequence data supporting the findings of this work are accessible through GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) with the accession number MH706708. The related BioProject, SRA, and Bio-Sample identifiers are PRJNA790665, SRP351584, and SAMN24219952.
Additional information
Funding
References
- Aguirre-Planter É, Jaramillo-Correa JP, Gómez-Acevedo S, Khasa DP, Bousquet J, Eguiarte LE. 2012. Phylogeny, diversification rates and species boundaries of Mesoamerican firs (Abies, Pinaceae) in a genus‐wide context. Mol Phylogenet Evol. 62(1):263–274.
- Blazier JC, Guisinger MM, Jansen RK. 2011. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol Biol. 76(3–5):263–272.
- Christian T. 2021. Abies salouenensis.Trees and Shrubs Online. [Accessed 2022 Dec 27]. treesandshrubsonline.org/articles/abies/abies-salouenensis/.
- Dallimore W, Jackson AB. 1966. A handbook of Coniferae and Ginkgoceae. 4th ed. London: Edward Arnold (Publishers) Ltd.
- Dong NL, Wang W, Wang Z, Zhang YY, Shao YZ, Sun HZ. 2021. Characterization of the complete plastid genome of Abies forrestii (Pinaceae) from southwest China. Mitochondrial DNA B Resour. 6(9):2772–2774.
- Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, Cheng T, Guo J, Zhou S. 2015. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 5:8348.
- Farjon A. 1990. Pinaceae: drawings and descriptions of the genera Abies, Cedrus, Pseudolarix, Keteleeria, Nothotsuga, Tsuga, Cathaya, Pseudotsuga, Larix and Picea (Regnum Vegetabile 121). Königstein: Koeltz Scientific Books; p. 9–109.
- Farjon A. 2001. World checklist and bibliography of conifers. 2nd ed. London: RoyalBotanic Gardens, Kew; p. 109–134.
- Farjon A, Rushforth KD. 1989. A classification of Abies Miller (Pinaceae). Notes Roy Bot Gard Edinburgh. 46(1):59–77.
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32:W273–W279.
- Fu ZX, Wang XY, Fan PZ, Tian XY, Shao YZ. 2019. The complete chloroplast genome of the endangered Pinaceae species Abies ziyuanensis and its phylogenetic implications. Mitochondrial DNA B. 4(1):137–138.
- Guo ZN, Lu XF, Dong YB, Wu JC, Li GY. 2019. Next-generation sequencing yields the complete chloroplast genome of Abies fanjingshanensis, an endangered species from South China. Mitochondrial DNA B. 4(1):880–881.
- Handel-Mazzetti H. 1929. Symbolae sincae. Vol. 7. Wien: J. Springer; p. 4–18.
- Li JJ, Cheng NY, Shi YC. 2021. The complete chloroplast genome of Keteleeria davidiana var. calcarea (Pinaceae), an endangered species endemic to China. Mitochondrial DNA B Resour. 6(2):693–695.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kaur S, Panesar PS, Bera MB, Kaur V. 2015. Simple sequence repeat markers in genetic divergence and marker-assisted selection of rice cultivars: a review. Crit Rev Food Sci Nutr. 55(1):41–49.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kuan CT. 1981. Fundamental features of the distribution of Coniferae in Sichuan. Acta Phytotaxonomica Sinica. 19(4):393–407.
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29(22):4633–4642.
- Liepelt S, Mayland-Quellhorst E, Lahme M, Ziegenhagen B. 2010. Contrasting geographical patterns of ancient and modern genetic lineages in Mediterranean Abies species. Plant Syst Evol. 284(3-4):141–151.
- Liu M-L, Bai J-Q, Dong W-L, Wang R-N, Dong P-B, Wang Ning, Liu H-Y, Fang M-F. 2018. Characterization of the whole plastid genome sequence of Abies chensiensis (Pinaceae), an endangered endemic conifer in China. Mitochondrial DNA B Resour. 3(2):1141–1142.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 0:1–11.
- Liu TS. 1971. A monograph of the genus Abies. Taipei: National Taiwan University, Department of Forestry, College of Agriculture.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:75–81.
- Rushforth KD. 1984. Notes on Chinese silver firs: 2. Notes Roy Bot Gard Edinburgh. 41:535–540.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan, and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Semerikova SA, Khrunyk YY, Lascoux M, Semerikov VL. 2018. From America to Eurasia: a multigenomes history of the genus Abies. Mol Phylogenet Evol. 125:14–28.
- Semerikova SA, Semerikov VL, Lascoux M. 2011. Post-glacial history and introgression in Abies (Pinaceae) species of the Russian Far East inferred from both nuclear and cytoplasmic markers. J Biogeogr. 38(2):326–340.
- Shao YZ, Chen Y, Lu XF, Ye YZ, Yuan ZL. 2019. Next-generation sequencing yields the complete chloroplast genome of Abies kawakamii. Mitochondrial DNA B Resour. 4(1):29–30.
- Shao YZ, Chen Y, Zhang XC, Xiang QP. 2020. Species delimitation and phylogeography of Abies delavayi complex: inferred from morphological, molecular, and climatic data. J Syst Evol. 58(3):234–246.
- Shao YZ, Hu JT, Fan PZ, Liu YY, Wang YH. 2018. The complete chloroplast genome sequence of Abies beshanzuensis, a highly endangered fir species from South China. Mitochondrial DNA B. 3(2):923–924.
- Shao YZ, Shi ZY, Wang Z, Wang W, Chen Y, Wen Q. 2022. The complete chloroplast genome of Abies ernestii Rehder (Pinaceae) and its phylogenetic implications. Mitochondrial DNA B Resour. 7(8):1497–1503.
- Shao YZ, Xiang QP. 2015. Species delimitation and phylogeography of Abies chensiensis complex: inferred from morphological and molecular data. Bot J Linn Soc. 177(2):175–188.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Su L, Zhao PF, Lu XF, Shao YZ. 2019. The complete chloroplast genome sequence of Abies chensiensis (Pinaceae). Mitochondrial DNA B Resour. 4(2):3262–3263.
- Suyama Y, Yoshimaru H, Tsumura Y. 2000. Molecular phylogenetic position of Japanese Abies (Pinaceae) based on chloroplast DNA sequences. Mol Phylogenet Evol. 16(2):271–277.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wu S, Lu XF, Zhao PF, Shao YZ. 2019. Next-generation sequencing yields the complete chloroplast genome of Abies balsamea. Mitochondrial DNA B. 4(1):1445–1446.
- Xiang QP, Wei R, Shao YZ, Yang ZY, Wang XQ, Zhang XC. 2015. Phylogenetic relationships, possible ancient hybridization, and biogeographic history of Abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes. Mol Phylogenet Evol. 82:1–14.
- Xiang QP, Wei R, Zhu YM, Harris AJ, Zhang XC. 2018. New infrageneric classification of Abies in light of molecular phylogeny and high diversity in western North America. J Syt Evol. 56(5):562–572.
- Xiang QP, Xiang QY, Guo YY, Zhang XC. 2009. Phylogeny of Abies (Pinaceae) inferred from nrITS sequence data. Taxon. 58(1):141–152.
- Xiang QP, Xiang QY, Liston A, Zhang XC. 2004. Phylogenetic relationships in Abies (Pinaceae): Evidence from PCR‐RFLP of the nuclear ribosomal DNA internal transcribed spacer region. Bot J Linn Soc. 145(4):425–435.
- Yi DK, Choi K, Joo M, Yang JC, Mustafina FU, Han J-S, Son DC, Chang KS, Shin CH, Lee Y-M. 2016. The complete chloroplast genome sequence of Abies nephrolepis (Pinaceae: abietoideae). J Asia Pac Biodivers. 9(2):245–249.
- Yi DK, Yang JC, So S, Joo MJ, Kim DK, Shin CH, Lee YM, Choi K. 2015. The complete plastid genome sequence of Abies koreana (Pinaceae: abietoideae). Mitochondrial DNA. 27(4):1.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhang YY, Xiang RC, Dong NL, Liu YY, Shao YZ. 2020. Next-generation sequencing yields the complete chloroplast genome of Abies yuanbaoshanensis, an endangered species from South China. Mitochondrial DNA B Resour. 5(4):3821–3822.
