Abstract
Calophyllum soulattri Burm. f. (1768) is an evergreen tree native to Southeast Asia, Australia, and the Solomon Islands. It is known for its medicinal uses and has been utilized in traditional folk medicine. However, genomic resources for this species are still unavailable. In this study, we sequenced and assembled the first complete chloroplast genome of C. soulattri using next-generation sequencing data. The chloroplast genome of C. soulattri is 161,381 bp in length with a total GC content of 36.36%. The chloroplast genome contains a large single copy (LSC) region of 88,680 bp, a small single copy (SSC) region of 17,453 bp, and two inverted repeat (IR) regions of 27,624 bp each. Furthermore, the chloroplast genome has 131 genes, which include 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis indicated that C. soulattri is clustered in the same branch with C. inophyllum and is closely related to Mesua ferrea.
Introduction
Calophyllum is a genus that belongs to the family Calophyllaceae under the Clusioid clade of the order Malpighiales (Angiosperm Phylogeny Group Citation2009). Calophyllum species are known for their use in traditional medicine. For example, oils extracted from Calophyllum seeds are used to treat wounds and infections (Léguillier et al. Citation2015). Furthermore, the genus Calophyllum is a potential source of compounds for drug discovery and development due to the presence of different secondary metabolites in plant tissues such as terpenes (Nigam et al. Citation1988), xanthones (Mah et al. Citation2015), coumarins (Ee et al. Citation2011), phenols, and flavonoids (Hapsari et al. Citation2022). In fact, coumarins extracted from a Calophyllum species were found to be potent against HIV-1 virus (Spino et al. Citation1998). Aside from being used in traditional medicine, Calophyllum species are also known for being sources of wood for locals (Rabena and Macandog Citation2017). Calophyllum soulattri Burm.f. (1768) is a small tree species with opposite leaves that are ovate to elliptical or sub-oblong; it has black or purple fruit and a white inflorescence (). Its distribution is limited to mainland Southeast Asia, the Solomon Islands, and northern Australia (Pelser et al. Citation2011). However, the complete chloroplast genome sequence of C. soulattri has not been reported. This study reports the first complete chloroplast genome sequence of C. soulattri that provides a reference for understanding the phylogenetic relationship and plastome evolution in the Calophyllaceae and the clusioid clade.
Materials
Plant Materials
Disease-free leaf samples of C. soulattri were collected from the germplasm collection of the Metallophytes Laboratory, Forest Biological Sciences, College of Forestry and Natural Resources, University of the Philippines, Los Baños, Laguna, Philippines (14°9′17″N 121°14′6.25″E). The leaf specimen was submitted at the Jose Vera Santos Memorial Herbarium of the Institute of Biology, University of the Philippines, Diliman (https://biology.science.upd.edu.ph/index.php/jose-vera-santos-memorial-herbarium/, Dr. Edwino S. Fernando, [email protected]; and compared with accession number 14288 (). Leaves were thoroughly cleaned and flash frozen using liquid nitrogen and were brought to the Plant Molecular Phylogenetics Laboratory at the Institute of Biology, University of the Philippines, Diliman, Quezon City for DNA extraction.
Methods
The total genomic DNA of C. soulattri was extracted using Wizard® HMW DNA Extraction Kit (Promega, USA) with slight modification by adding 1% (w/v) polyvinylpyrrolidone-40 (PVP-40) to the HMW lysis buffer A and submitted to MACROGEN, Inc. (Seoul, South Korea) for next-generation sequencing service. Paired-end reads were sequenced using the Illumina HiSeq 2500 platform (San Diego, California). Approximately, 2.8 Gb of raw data was generated. The quality of raw reads was checked using FastQC v0.11.9 (Andrews Citation2010) and read error correction was performed using Musket v1.1 (Liu et al. Citation2013). The plastome of C. soulattri was assembled using GetOrganelle v1.7.6.1 (Jin et al. Citation2020). Subsequently, the assembled chloroplast genome was annotated using GeSeq (Tillich et al. Citation2017) and then manually curated. The chloroplast genome map was drawn using the CPGView program (Liu et al. Citation2023).
To validate the phylogenetic position of C. soulattri, the complete chloroplast genome sequences of 22 species in the order Malpighiales were downloaded from NCBI GenBank and aligned using MAFFT v7.490 (Katoh and Standley Citation2013) on the CIPRES Science Gateway (www.phylo.org) (Miller et al. Citation2015). The maximum-likelihood (ML) phylogenetic analysis was performed using RaxML-NG v1.1.0 (Kozlov et al. Citation2019) based on GTR + G nucleotide substitution model on the CIPRES Science Gateway (www.phylo.org) (Miller et al. Citation2015) with 1,000 bootstrap replicates and Bayesian inference using MrBayes v3.2.7a (Ronquist et al. Citation2012). Lophira alata (MZ274135) (Mascarello et al. Citation2021) from the Ochnaceae family and Passiflora edulis (NC_034285) (Cauz-Santos et al. Citation2017) was used as an outgroup.
Results
General characteristic of the chloroplast genome
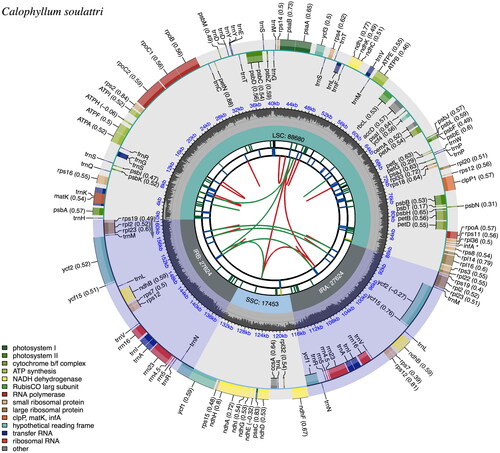
The complete chloroplast genome (plastome) of C. Soulattri has a typical circular and quadripartite structure. The plastome is 161,381 bp in length with a total GC content of 36.36% consisting of the large single copy (LSC: 88,680 bp) and small single copy (SSC: 17,453 bp) regions, which are separated by two inverted repeat regions (IRa and IRb: 27,624 bp each). The GC contents of the LSC, SSC, and IR regions were 33.96%, 30.57%, and 42.04%, respectively (). The average and minimum read mapping depth of the assembled plastome were 97.3x and 80.3x, respectively (Figure S1). Furthermore, the plastome of C. soulattri has 131 genes including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
Figure 2. The chloroplast genome (plastome) map of C. soulattri. The plastome map is divided into six circles with different representations. Starting from the inner circle, the first circle shows the dispersed repeats connected by red (forward repeat) and green (palindromic repeat) arcs. The second circle shows long tandem repeats in short blue bars. The third circle presents simple sequence repeats (SSRs) that are color coded based on repeat unit size (RUS) (Black, complex repeat; Green, RUS = 1; Yellow, RUS = 2; and Blue, RUS = 4). The fourth circle displays the four regions of the plastome (LSC, SSC, IRa, and IRb) with their respective sizes. The fifth circle provides the GC content along the genome. The sixth circle provides the genes with their codon usage bias in parentheses and are color-coded, indicating their respective functional group. The functional groups can be found at the bottom left corner. Genes that are found in the inner and outer circle are transcribed clockwise and counterclockwise, respectively.
Phylogenetic analysis of C. soulattri in the Clusioid clade
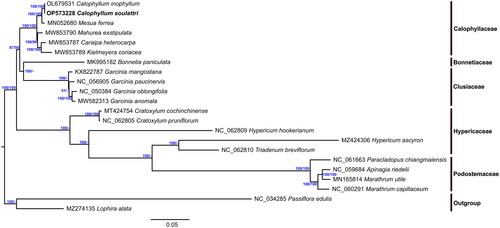
Our ML phylogenetic analysis revealed two major clades with high bootstrap support (BS = 100%) (). Garcinia (Clusiaceae) formed a monophyletic clade with a highly supported bootstrap value (BS = 100%). The family Hypericaceae was related to Podostemaceae. Moreover, the family Calophyllaceae also formed a monophyletic clade (BS = 100%) in which M. ferrea was related to C. soulattri and C. inophyllum indicating that they are more closely related than to other species.
Figure 3. Maximum likelihood (ML) phylogenetic tree and Bayesian inference tree of C. soulattri and other Clusioid species from the order Malpighiales based on the whole chloroplast genome sequence with L. alata and P. edulis as outgroup. Values on the right represents bootstrap support from Maximum Likelihood analysis while on the left represents Bayesian posterior probabilites. The species shown in bold font is newly sequenced in this study. The following sequences were used: C. inophyllum OL679531, M. ferrea MN052680 (Wang et al. Citation2019), Bonnetia paniculata MK995182 (Jin et al. Citation2020), Garcinia mangostana KX822787 (Jo et al. Citation2017), G. paucinervis MT501656 (Wang et al. Citation2021), G. oblongifolia MT726019 (Ma et al. Citation2020), G. anomala MW582313 (Yue and Shi Citation2021), Caraipa exstipulata MW853790 (Trad et al. Citation2021), Kielmeyera coriacea MW853789 (Trad et al. Citation2021), C. heterocarpa MW853787 (Trad et al. Citation2021), Apinagia riedelii MN165812 (Bedoya et al. Citation2019), Marathrum utile MN165814 (Bedoya et al. Citation2020), M. capillaceum MN165813 (Bedoya et al. Citation2020), Paracladopus chiangmaiensis MZ645928 (Wu et al. Citation2022), Hypericum ascyron MZ424306 (Claude et al. Citation2022), Triadenum breviflorum MZ714016 (Sudmoon et al. Citation2022), H. hookerianum MZ714015 (Sudmoon et al. Citation2022), Cratoxylum pruniflorum MZ703416 (Sudmoon et al. Citation2022), C. cochinchinense MT424754 (Huang et al. Citation2019), Lophira alata MZ274135 (Mascarello et al. Citation2021), and NC_034285 Passiflora edulis (Cauz-Santos et al. Citation2017). NCBI accession numbers are given for Genbank sequences.
Discussion and conclusion
The first complete chloroplast genome (plastome) of C. soulattri was sequenced, assembled, and annotated in this study. The general features of the chloroplast genome of C. soulattri are similar to that of most land plants. Furthermore, our results mostly concur with a recent study which elucidates a generally conserve plastome structure (Trad et al. Citation2021).
Calophyllum soulattri belongs to the tribe Calophylleae of the family Calophyllaceae. There are almost 90 genera and approximately 2,090 species within the family Calophyllaceae. Also, the family Calophyllaceae is monophyletic along with Clusiaceae, Bonnetiacea, Podostemaceae, and Hypericaceae in the order Malpighiales which forms a group typically known as the clusiod clade (Stevens Citation2001 onwards; Cook and Rutishauser Citation2007; Angiosperm Phylogeny Group Citation2009; Wurdack and Davis Citation2009; Ruhfel et al. Citation2011; Koi et al. Citation2012; Nürk et al. Citation2013). Although the group is species-rich, complete plastome information is still unavailable for most species.
Our phylogenetic analysis using available whole chloroplast genomes has suggested a different topology with high bootstrap support than those previously reported. Nevertheless, species groupings are consistent in the clusioid clade as has been suggested in several studies (Ruhfel et al. Citation2011; Citation2016; Trad et al. Citation2021). Furthermore, previous reports support our findings on M. ferrea being closely related to Calophyllum species using plastid, mitochondrial, and nuclear sequences, and even the whole chloroplast genome (Ruhfel et al. Citation2016; Cabral et al. Citation2021; Trad et al. Citation2021). This study provides significant data for understanding species placement in the clusioid clade which would be useful in future analyses. Thus, the complete chloroplast genome of C. soulattri provides a valuable genetic resource for future studies on the phylogeny and population genetics of Calophyllum.
Ethical approval
This project has obtained permission to collect plant samples through the Department of Natural Resources with DENR BMB Wildlife Gratuitous Permit No. 299, issued Sept 2019. The plant material collection and experimental research were conducted according to RA 9147 or the Wildlife Resources Conservation and Protection Act of the Philippines.
Authors’ contributions
JRey was involved in conceptualization and design, analysis, and interpretation of the data; revising it critically for intellectual content; and the final approval of the version to be published while DPahayo and CACadorna were involved in the design, analysis, and interpretation of the data; the drafting of the paper and revising it critically for intellectual content; MOQuimado provided plant tissue samples and drafting and finalization of manuscript and that all four authors agree to be accountable for all aspects of the work.
Supplemental Material
Download PDF (258.4 KB)Supplemental Material
Download JPEG Image (925.8 KB)Supplemental Material
Download JPEG Image (905.3 KB)Supplemental Material
Download PNG Image (951.9 KB)Supplemental Material
Download JPEG Image (553.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The chloroplast genome sequence data that support the findings of this study are openly available in GenBank of NCBI database at (https://www.ncbi.nlm.nih.gov/) under the accession no. OP573228. The associated BioProject, SRA, and Biosample numbers of C. soulattri are PRJNA891016, SRR22031315, and SAMN31427486, respectively.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data [accessed 2022 Sep 10]. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Angiosperm Phylogeny Group. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161(2):105–121.
- Bedoya AM, Ruhfel BR, Philbrick CT, Madriñán S, Bove CP, Mesterházy A, Olmstead RG. 2019. Plastid genomes of five species of Riverweeds (Podostemaceae): structural organization and comparative analysis in Malpighiales. Front Plant Sci. 10:1035.
- Bedoya AM, Ruhfel BR, Philbrick CT, Madriñán S, Bove CP, Mesterházy A, Olmstead RG. 2020. Corrigendum: plastid genomes of five species of Riverweeds (Podostemaceae): structural organization and comparative analysis in Malpighiales. Front Plant Sci. 11:598458.
- Cabral FN, Trad RJ, Amorim BS, Maciel JR, do Amaral MDCE, Stevens P. 2021. Phylogeny, divergence times, and diversification in Calophyllaceae: linking key characters and habitat changes to the evolution of Neotropical Calophylleae. Mol Phylogenet Evol. 157:107041.
- Cauz-Santos LA, Munhoz CF, Rodde N, Cauet S, Santos AA, Penha HA, Dornelas MC, Varani AM, Oliveira GCX, Bergès H, et al. 2017. The chloroplast genome of Passiflora edulis (Passifloraceae) assembled from long sequence reads: structural organization and phylogenomic studies in Malpighiales. Front Plant Sci. 8:334.
- Claude SJ, Park S, Park S. 2022. Gene loss, genome rearrangement, and accelerated substitution rates in plastid genome of Hypericum ascyron (Hypericaceae). BMC Plant Biol. 22(1):1–12.
- Cook CDK, Rutishauser R. 2007. Podostemaceae. In: Kubitzki K, editors. Flowering Plants · Eudicots. The Families and Genera of Vascular Plants. Springer. 9: 304–344
- Ee GCL, Mah SH, Teh SS, Rahmani M, Go R, Taufiq–Yap YH. 2011. Soulamarin, a new coumarin from stem bark of Calophyllum soulattri. Molecules. 16(11):9721–9727.
- Hapsari S, Yohed I, Kristianita RA, Jadid N, Aparamarta HW, Gunawan S. 2022. Phenolic and flavonoid compounds extraction from Calophyllum inophyllum leaves. Arab J Chem. 15(3):103666.
- Huang J, Wang Y, Xu S, He J, Zhang Z. 2019. The complete chloroplast genome of Cratoxylum cochinchinense (Hypericaceae). Mitochondrial DNA Part B Resour. 4(2):3452–3453.
- Jin DM, Jin JJ, Yi TS. 2020. Plastome structural conservation and evolution in the clusioid clade of Malpighiales. Sci Rep. 10(1):9091.
- Jin JJ, Yu WB, Yang JB, Song Y, De Pamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Jo S, Kim HW, Kim YK, Sohn JY, Cheon SH, Kim KJ. 2017. The complete plastome of tropical fruit Garcinia mangostana (Clusiaceae). Mitochondrial DNA Part B Resour. 2(2):722–724.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Koi S, Kita Y, Hirayama Y, Rutishauser R, Huber KA, Kato M. 2012. Molecular phylogenetic analysis of Podostemaceae: implications for taxonomy of major groups. Bot J Linn Soc. 169(3):461–492.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML–NG: a fast, scalable and user–friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Léguillier T, Lecsö-Bornet M, Lémus C, Rousseau-Ralliard D, Lebouvier N, Hnawia E, Nour M, Aalbersberg W, Ghazi K, Raharivelomanana P, et al. 2015. The wound healing and antibacterial activity of five ethnomedical Calophyllum inophyllum oils: an alternative therapeutic strategy to treat infected wounds. PLOS One. 10(9):e0138602.
- Liu Y, Schröder J, Schmidt B. 2013. Musket: a multistage k–mer spectrum–based error corrector for Illumina sequence data. Bioinformatics. 29(3):308–315.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 0:1–11.
- Ma X, Chen W, Tang L. 2020. The complete chloroplast genome sequence of Garcinia oblongifolia (Clusiaceae). Mitochondrial DNA Part B Resour. 5(3):3206–3207.
- Mah SH, Ee GCL, Teh SS, Sukari MA. 2015. Antiproliferative xanthone derivatives from Calophyllum inophyllum and Calophyllum soulattri. Pak J Pharm Sci. 28(2):425–430.
- Mascarello M, Amalfi M, Asselman P, Smets E, Hardy OJ, Beeckman H, Janssens SB. 2021. Genome skimming reveals novel plastid markers for the molecular identification of illegally logged African timber species. PLOS One. 16(6):e0251655.
- Miller MA, Schwartz T, Pickett BE, He S, Klem EB, Scheuermann RH, Passarotti M, Kaufman S, O'Leary MA. 2015. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol Bioinform Online. 11:43–48.
- Nigam SK, Banerji R, Rebuffat S, Cesario M, Pascard C, Bodo B. 1988. Soulattrone A, A C24 terpenoid from Calophyllum soulattri. Phytochemistry. 27(2):527–530.
- Nürk NM, Madriñán S, Carine MA, Chase MW, Blattner FR. 2013. Molecular phylogenetics and morphological evolution of St. John’s wort (Hypericum; Hypericaceae). Mol Phylogenet Evol. 66(1):1–16.
- Pelser PB, Barcelona JF, Nickrent DL (eds). 2011 onwards. Co’s Digital Flora of the Philippines; [accessed 2022 Sep 1]. https://www.philippineplants.org.
- Rabena MAF, Macandog DM. 2017. Contemporary knowledge of woodlot (Muyong) resource management: a case study of key–informants’ perceptions in Brgy. Kinakin, Banaue, Ifugao, Philippines. J Manag Dev Stud. 6:14–21.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Ruhfel BR, Bittrich V, Bove CP, Gustafsson MHG, Philbrick CT, Rutishauser R, Xi Z, Davis CC. 2011. Phylogeny of the clusioid clade (Malpighiales): evidence from the plastid and mitochondrial genomes. Am J Bot. 98(2):306–325.
- Ruhfel BR, Bove CP, Philbrick CT, Davis CC. 2016. Dispersal largely explains the Gondwanan distribution of the ancient tropical clusioid plant clade. Am J Bot. 103(6):1117–1128.
- Spino C, Dodier M, Sotheeswaran S. 1998. Anti–HIV coumarins from Calophyllum seed oil. Bioorg Med Chem Lett. 8(24):3475–3478.
- Stevens PF. 2001. onwards. Angiosperm Phylogeny Website. Missouri Botanical Garden [accessed 2022 Sep 15]. https://www.mobot.org/MOBOT/research/APweb/.
- Sudmoon R, Kaewdaungdee S, Tanee T, Siripiyasing P, Ameamsri U, Syazwan SA, Lee SY, Chaveerach A. 2022. Characterization of the plastid genome of Cratoxylum species (Hypericaceae) and new insights into phylogenetic relationships. Sci Rep. 12(1):1–11.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht–Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Trad RJ, Cabral FN, Bittrich V, Silva SRD, Amaral M. 2021. Calophyllaceae plastomes, their structure and insights in relationships within the clusioids. Sci Rep. 11(1):1–15.
- Wang Y, Yuan X, Li Y, Zhang J. 2019. Complete chloroplast genome of Mesua ferrea: the first Calophyllaceae plastome. Mitochondrial DNA Part B Resour. 4(2):3027–3028.
- Wang Y, Zhao B, Lu Z, Shi Y, Li J. 2021. The complete chloroplast genome provides insight into the polymorphism and adaptive evolution of Garcinia paucinervis. Biotechnol Biotechnol Equip. 35(1):377–391.
- Wick R, Schultz M, Zobel J, Holt K. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Wu M, Zhang K, Yang X, Qian X, Li R, Wei J. 2022. Paracladopus chiangmaiensis (Podostemaceae), a new generic record for China and its complete plastid genome. PhytoKeys. 195:1–13.
- Wurdack KJ, Davis CC. 2009. Malpighiales phylogenetics: gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am J Bot. 96(8):1551–1570.
- Yue B, Shi J. 2021. The complete chloroplast genome sequence of Garcinia anomala (Clusiaceae) from Yunnan Province, China. Mitochondrial DNA Part B Resour. 6(7):1899–1900.

