Abstract
The VIOLA ALBIDA complex is a complicated group with taxonomic problems having continuous leaf variations and composed of taxa related to the following names: Viola albida, V. albida var. takahashii, and V. chaerophylloides. As a first step to understanding the genomic nature of this complex, this study identified the whole chloroplast genome of V. albida. The genome is 157,692 bp in length (36.3% of GC content) and contains four subregions: a large single copy region of 86,220 bp, a small single copy region of 17,248 bp, and a pair of inverted regions of 27,112 bp each. An annotation of the gene identifies 111 unique genes, including 77 protein-coding genes, four rRNA genes, and 30 tRNA genes. The phylogenetic analysis of this genome with selected cp genomes from Viola identifies the close relationship between V. albida and V. ulleungdoensis. It is noteworthy that V. chaerophylloides, traditionally recognized as a member of the VIOLA ALBIDA complex, is genetically distant from V. albida and forms a sister group of all other members of the subsection Patellares. Our genome report is expected to serve as a basis for understanding the identity of the VIOLA ALBIDA complex.
Introduction
Viola L. (Violaceae) is the 40th to 50th largest genera in angiosperms and contains ca. 664 species mainly distributed in temperate regions and highlands in tropical mountains (Marcussen et al. Citation2022). The type specimen of Viola albida Palibin (Palibin Citation1899) was collected in Seoul, Korea, and the species is distributed in far Eastern Russia, Northern China, and the Korean peninsula (https://powo.science.kew.org/). The species is included in subgenus Viola, section Plagiostigma, and subsection Patellares in the recent classification system of Viola (Marcussen et al. Citation2022). V. albida is distinguished from other Viola species in Korea by plants without stems, showy white flowers having spherical-apex style, stipules adnate to petiole, undissected leaves having shallowly serrated margins, etc. (Lee and Yoo Citation2020, ). As closely related taxa, V. albida, V. albida var. takahashii (Nakai) Nakai (Nakai Citation1916), and V. chaerophylloides (Regel) W. Becker (Becker Citation1902) exhibit morphological continuity in leaves: from undivided (V. albida) through deeply lobed (V. albida var. takahashii) to fully divided (compound leaves; V. chaerophylloides) (Choi and Whang Citation2019). Since they have a complex taxonomic history, often recognized as independent species or varieties of V. albida, these taxa are recognized as the VIOLA ALBIDA complex (Whang Citation2006; Choi and Whang Citation2019). Various approaches have been conducted to understand the evolution of VIOLA ALBIDA complex: leaf morphological (Kim et al. Citation1991; Jang et al. Citation2006), genetic (Ko et al. Citation1998; Koo et al. Citation2010), developmental (Choi and Whang Citation2019), transcriptome (Srikanth et al. Citation2019), and phylogenetic (Whang Citation2006) studies. Interestingly, two previous molecular phylogenetic studies of Korean violets based on two (Yoo et al. Citation2007) and eight (Yoo and Jang Citation2010) chloroplast (cp) DNA regions, respectively, showed that VIOLA ALBIDA complex do not form an independent clade: these studies provided insufficient phylogenetic information. Therefore, unraveling the phylogenomic structure of the VIOLA ALBIDA complex requires analyses with more data, such as analyses based on cp whole-genome data or nuclear HybSeq (Weitemier et al. Citation2014) data. As the first step for understanding the genome evolution of the VIOLA ALBIDA complex, we report a complete chloroplast genome sequence of V. albida, a typical member of the VIOLA ALBIDA complex, in this study.
Materials and methods
A sample for the study was collected from Mt. Bukhan, Seoul, Korea (N 37°37'47.04", E 126°58'35.89"). The voucher specimen (S. Kim 2020-001, SWU) was deposited at the herbarium of Sungshin Women’s University (SWU; Sangtae Kim, [email protected]). With a commercial kit (ExgeneTM Plant SV; GeneAll Biotechnology Co. Ltd., Seoul, Korea), total genomic DNA was extracted from a fresh leaf. We produced 25,132,239 paired-end reads (150 bp) using the MGISEQ-2000 platform (MGI Tech Co. Ltd., Shenzhen, China). Following the manufacturer’s manual, a paired-end sequencing library was prepared with the MGIEasy DNA library Prep kit (MGI Tech Co. Ltd., Shenzhen, China). After the library construction, the extracted DNA was stored in the DNA Bank of the Plant Molecular Phylogeny Lab, Department of Biology, Sungshin Women’s University. We obtained 7.53 Gbps of sequence after the low-quality filtration (>Q30: 88.6%) by Trimmomatic (Bolger et al. Citation2014). In order to determine the cp genome of V. albida, the paired-end reads were mapped against the cp genome of V. ulleungdoensis (NC_050744), of which phylogenetic position is closely related to the members of the VIOLA ALBIDA complex (Yoo and Jang Citation2010; Yang et al. Citation2021), with the mapping module in the Geneious prime (v. 11.0.11 + 7; 'medium-low’ sensitivity option; Kearse et al. Citation2012). When we manually checked the entire sequence regions, the whole genome was perfectly mapped with apparent substitutions and indels, and no severe mismatching regions were detected. A consensus sequence was produced, and it was confirmed again with the mapping of raw reads against the consensus sequence for the completion of cp genome of V. albida. Gene annotation of the consensus sequence was performed by GeSeq (Tillich et al. Citation2017) with the seven previously reported Viola cp genomes (NC_050744.1, NC_041585.1, NC_026986.1, NC_041584.1, NC_052919.1, NC_041583.1, and NC_041582.1) as references. The annotations were manually checked and edited by the comparison with the above seven Viola cp genomes using Geneious prime (v. 11.0.11 + 7; Kearse et al. Citation2012). The chloroplast genome map of V. albida was drawn with OGDRAW (Greiner et al. Citation2019). The structure of the cis-splicing genes (Figure S1A) and the structure of the trans-splicing gene rps12 (Figure S1B) in the chloroplast genome of V. albida were recognized using CPGview (Liu et al. Citation2023).
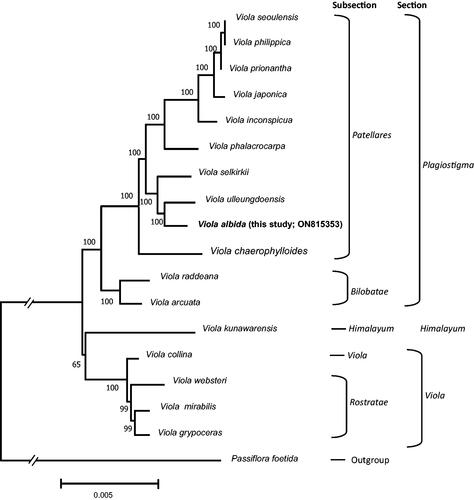
To show the phylogenetic position of V. albida in Viola, we analyzed 16 previously reported complete cp genomes in Violaceae, including V. albida, and an outgroup (Passiflora foetida; Passifloraceae). The selection of an outgroup for the phylogenetic analysis was based on a recent phylogenetic study (Xi et al. Citation2012). The genome sequences were aligned using MAFFT (v7.450; Katoh and Standley Citation2013) with the default option. For the Maximum-Likelihood (ML) analysis, the best model (General Time Reversible + G + I) was selected by the model test module included in the MEGAX (Kumar et al. Citation2018). For the ‘Gaps/Missing Data Treatment’, 95% of the site coverage cutoff was applied in both the model test and ML analysis. Five hundred bootstrap replications were applied to address node reliability in the ML tree.
Results
The cp genome sequence of V. albida was completed with an average coverage of 3,810.51 X (Figure S2). The complete cp genome of V. albida (GenBank accession number: ON815353) is 157,692 bp in length (36.3% of GC content) and consists of an LSC region of 86,220 bp, an SSC region of 17,248 bp, and a pair of IR regions of each 27,112 bp (). There are 111 unique genes in the cp genome of V. albida, including 77 protein-coding genes, four rRNA genes, and 30 tRNA genes. The phylogenetic tree () shows that V. albida, of which the cp genome is determined in this study (ON815353), forms a clade with V. ulleungdoensis, and this clade is a sister to V. selkirkii. When we compare cp genome sequences between V. albida and V. ulleungdoensis, 647 substitutions, and 1,050 indels were identified. V. chaerophylloides is a sister to all other members of the subsection Patellares. The bootstrap analysis supports all of these relationships with high bootstrap values.
Figure 2. The circular chloroplast genome map of V. albida drawn by OGDRAW. Arrows indicate the direction of transcription of genes located inside and outside the large circle. The histogram inside the small circle shows the GC contents. Asterisk (*) indicates genes containing introns.
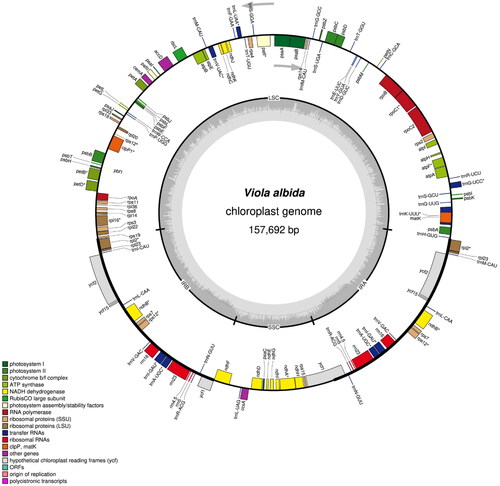
Figure 3. A tree generated from the maximum-likelihood analysis with 17 selected Viola cp genomes, including a genome determined in this study (V. albida) and an outgroup (Passiflora foetida). The number above the node indicates the bootstrap support with 500 replications. The following sequences were used: V. seoulensis NC_026986 (Cheon et al. Citation2017), V. philippica NC_052919 (Guo et al. Citation2021), V. prionantha NC_057663 (Duan et al. Citation2020), V. japonica NC_057486 (Cheon et al. Citation2020), V. inconspicua MW802532 (Cao et al. Citation2022), V. phalacrocarpa NC_041583 (Cheon et al. Citation2019), V. selkirkii NC_059909 (unpublished), V. ulleungdoensis NC_050744 (Yang et al. Citation2021), V. albida ON815353 (this study), V. chaerophylloides NC_065000 (Cao et al. Citation2022), V. raddeana NC_041584 (Cheon et al. Citation2019), V. arcuata NC_061694 (Moon and Kim Citation2022), V. kunawarensis NC_060866 (Zhou et al. Citation2022), V. collina MW802530 (Cao et al. Citation2022), V. websteri NC_041585 (Cheon et al. Citation2019), V. mirabilis NC_041582 (Cheon et al. Citation2019), V. grypoceras NC_061911 (Park et al. Citation2023), and Passiflora foetida NC_043825 (Shrestha et al. Citation2019).
Discussion and conclusion
When determining chloroplast genome sequences using mapping methods, we can minimize errors using the genetically closest genome. However, previous molecular phylogenetic studies of Viola didn’t provide information for the most relative genome to V. albida (Yoo and Jang Citation2010; Yang et al. Citation2021) because trees from these studies showed polytomy or low bootstrap supports. We, therefore, further mapped the raw reads against the three candidate cp genomes [V. selkirkii (NC_059909), V. chaerophylloides (NC_065000), and V. phalacrocarpa (NC_041583)], respectively. The resulting sequences are all identical, supporting the reliability of the final cp sequence from V. albida.
A previous phylogenetic analysis using eight chloroplast DNA regions showed that the two members of the VIOLA ALBIDA complex, V. albida and V. chaerophylloides, do not form a monophyletic clade (Yoo and Jang Citation2010). Although our analysis did not include V. albida var. takahashii because its cp genome is not published yet, our analysis based on complete cp genomes also confirmed this. However, since our phylogenetic tree shows only maternal inheritance, this does not entirely rule out the possibility that the species belonging to the VIOLA ALBIDA complex is a monophyletic group. To clarify the taxonomic structure of this complex, future studies should include intensive nuclear data analysis.
In this study, we report the cp genome sequence of V. albida. The results of our study will provide essential information for future intensive phylogenomic and evolutionary studies of the VIOLA ALBIDA complex as well as the genus Viola.
Ethical approval
This research complies with Sungshin Women’s University Research Ethics Guidelines and related institutional, national, and international guidelines and laws. Since V. albida is a common plant in Korea and is not registered as a threatened or endangered species, a sampling permit is not required.
Author contributions
S. Kim organized this research, collected the sample, produced raw data, and revised the manuscript. H. Moon analyzed data and prepared a preliminary manuscript. All researchers agreed on the final manuscript.
Supplemental Material
Download MS Word (222.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. ON815353. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA875227, SRR21363874, and SAMN30608856, respectively.
Additional information
Funding
References
- Becker W. 1902. Ergebnisse einer revision der Violae des herbarium Barbey-Boissier. Bull Herb Boiss. Ser. 2:856.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Cao DL, Zhang XJ, Xie SQ, Fan SJ, Qu XJ. 2022. Application of chloroplast genome in the identification of Traditional Chinese Medicine Viola philippica. BMC Genomics. 23(1):540.
- Cheon KS, Kim DK, Kim KA, Yoo KO. 2020. The complete chloroplast genome sequence of Viola japonica (Violaceae). Mitochondrial DNA B Resour. 5(2):1297–1298.
- Cheon KS, Kim KA, Kwak M, Lee B, Yoo KO. 2019. The complete chloroplast genome sequences of four Viola species (Violaceae) and comparative analyses with its congeneric species. PLoS One. 14(3):e0214162.
- Cheon KS, Yang JC, Kim KA, Jang SK, Yoo KO. 2017. The first complete chloroplast genome sequence from Violaceae (Viola seoulensis). Mitochondrial DNA A: DNA Mapp. Seq. Anal. 28(1):67–68.
- Choi YK, Whang SS. 2019. A comparative study of early leaf development in the Viola albida complex. Korean J Pl Taxon. 49(1):1–7.
- Duan C, Zhang K, Duan YZ. 2020. The complete chloroplast genome sequence of Viola prionantha (Violaceae). Mitochondrial DNA B Resour. 5(3):2924–2926.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Guo Y, Lin P, Wang M. 2021. The complete chloroplast genome of Viola philippica. Mitochondrial DNA B Resour. 6(4):1494–1495.
- Jang SK, Lee WT, Yoo KO. 2006. Taxonomic study on Viola albida var. albida and its related taxa. Korean J Pl Taxon. 36(3):163–187.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim KS, Sun BY, Whang SS, Chung GH. 1991. Biosystematic study on the genus Viola in Korea -comparative morphology of the Viola albida complex. Korean J Bot. 34(3):229–238.
- Ko MK, Yang J, Jin YH, Lee CH, Oh BJ. 1998. Genetic relationships of Viola species evaluated by random amplified polymorphic DNA analysis. J Hortic Sci Biotechnol. 73(5):601–605.
- Koo JC, Tak HJ, Whang SS. 2010. Taxonomic study of Viola albida complex based on RAPD data. Korean J Pl Taxon. 40(2):118–129.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee WT, Yoo KO. 2020. Violaceae batsch. In: Park C, editor. Flora of Korea. Incheon: National Institute of Biological Resources. p. 45–59.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Marcussen T, Ballard HE, Danihelka J, Flores AR, Nicola MV, Watson JM. 2022. A revised phylogenetic classification for Viola (Violaceae). Plants. 11(17):2224.
- Moon H, Kim S. 2022. A chloroplast genome sequence of Viola arcuata distributed in Korea. Mitochondrial DNA B Resour. 7(9):1636–1638.
- Nakai T. 1916. Notulae ad plantas Japaoniae et Coreae. XII. Bot Mag. 30:286.
- Palibin J. 1899. Conspectus florae Koreae. Trudy Imp S -Peterburgsk Bot Sada. 17(1):30–31.
- Park JH, Lee M, Lee Y, Lee J. 2023. The complete chloroplast genome of Viola grypoceras (Violaceae). Mitochondrial DNA B Resour. 8(1):42–44.
- Shrestha B, Weng ML, Theriot EC, Gilbert LE, Ruhlman TA, Krosnick SE, Jansen RK. 2019. Highly accelerated rates of genomic rearrangements and nucleotide substitutions in plastid genomes of Passiflora subgenus Decaloba. Mol Phylogenet Evol. 138:53–64.
- Srikanth K, Hill RS, Whang SS. 2019. A correlation between leaf shape and its related key genes in Viola albida complex. In Vitro CellDevBiol-Plant. 55(4):409–420.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Weitemier K, Straub SCK, Cronn RC, Fishbein M, Schmickl R, McDonnell A, Liston A. 2014. Hyb-Seq: combining target enrichment and genome skimming for plant phylogenomics. Appl Plant Sci. 2(9):1400042.
- Whang SS. 2006. Analysis of ITS DNA sequences of the Viola albida complex. Korean J Plant Res. 19(5):628–633.
- Xi Z, Ruhfel BR, Schaefer H, Amorim AM, Sugumaran M, Wurdack KJ, Endress PK, Matthews ML, Stevens PF, Mathews S, et al. 2012. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci U S A. 109(43):17519–17524.
- Yang J, Park S, Gil HY, Park JH, Kim SC. 2021. Characterization and dynamics of intracellular gene transfer in plastid genomes of Viola (Violaceae) and order Malpighiales. Fron Plant Sci. 12:678580.
- Yoo KO, Jang SK. 2010. Infrageneric relationships of Korean Viola based on eight chloroplast markers. J Syst Evol. 48(6):474–481.
- Yoo KO, Jang SK, Lee WT. 2007. Phylogenetic relationships of Korean Viola (Violaceae) based on matK and atpB-rbcL sequence data of chloroplast DNA. Korean J Pl Taxon. 37(1):1–15.
- Zhou J, Zhang J, Qiu Y, Fan C. 2022. The complete chloroplast genome sequence of Viola kunawarensis Royle, a precious Uygur medicinal material. Mitochondrial DNA B Resour. 7(9):1704–1706.

