Abstract
The complete chloroplast genome sequence of Acer pseudosieboldianum (Sapindaceae) was determined. The chloroplast genome of A. pseudosieboldianum is 157,053 bp in length with two inverted repeats (26,747 bp) between a large single-copy (85,391 bp) and a small single-copy (18,168 bp). The GC content was 37.8% and it was composed of 86 coding genes, eight rRNA genes, 37 tRNA genes, and two pseudogenes, rps2, and ycf1. Molecular phylogenetic analysis based on the plastid genome sequences strongly supported the hypothesis that A. pseudosieboldianum was embedded in the series Palmata of section Palmata. However, the phylogenetic positions of A. ukurunduense and A. buergerianum, which are a members of the series Penninervia of sections Palmata and Pentaphylla, respectively, were incongruent with the recent sectional classification system.
Introduction
The genus Acer L. is a large plant group that includes over 120 species and is one of the most diverse and ecologically important tree genera in the northern temperate forest (Areces-Berazain et al. Citation2020; Yu et al. Citation2021). Although different infra-generic classification systems composed of 12–25 sections have been suggested, the relationships and circumscription in this group remain unclear (Suh et al. Citation2000; Grimm et al. Citation2006; Li et al. Citation2006). To resolve this problem, a phylogenomic study was recently conducted based on the complete chloroplast genome sequences (Areces-Berazain et al. Citation2020; Yu et al. Citation2021), using both genomic data and biogeographical information (Li et al. Citation2019) in the major lineages. Acer pseudosieboldianum (Pax) Kom. 1904, also known as a Korean maple or purplebloom maple, is taxonomically classified as a member of the largest section Palmata in the genus. In the present study, we confirmed the complete chloroplast genome sequence of A. pseudosieboldianum and evaluated its phylogenetic position within the genus.
Materials and methods
To obtain the complete chloroplast genome sequence of A. pseudosieboldianum, we collected plant material () from Jeongok-valley, Mt. Juwang-san, Korea (36° 36′ 92.57″ N, 129° 18′ 32.43″ E, alt. 338 m) and conducted the molecular phylogenetic analysis of the genus Acer using previously published genome data together. The voucher specimen of the plant material was deposited at the Herbarium of Kyungpook National University (KNU) under accession number Back-2016 ([email protected]). Total genomic DNA was extracted from a dehydrated leaf using silica gel by following the manufacturer’s protocol using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Then, the sample was sequenced using Illumina HiSeq4000 (Illumina, San Diego, CA) and totally 46,089,072 paired end reads (151 bp in length) were obtained. For trimming the raw data, we used trimmomatic 0.39 (Bolger et al. Citation2014) with the option LEADING:10, TRAILING:10, SLIDINGWINDOW:4:20, and MINLEN:50. After trimming, 622,944 reads were assembled into the reference plastome of A. palmatum (NC 034932) following Kim and Chase’s protocol (Kim and Chase Citation2017). The final circular cp genome sequence was constructed, and the coverage depth of the genome was 555.6 (Figure S1). Based on existing genes in other maple tree chloroplast genomes, all of the genes were annotated using Geneious 10.2.6 (Kearse et al. Citation2012) with manual correction and tRNAScan-SE (Lowe and Eddy Citation1997), especially for the tRNA gene. The cp genome map was drawn by CPGView program (Liu et al. Citation2023; http://www.1kmpg.cn/cpgview/). To understand the phylogenetic relationship of A. pseudosieboldianum and its related taxa, the phylogenetic tree was constructed based on the concatenated 78 coding genes of the released Sapindaceae chloroplast genome data using RAxML (Stamatakis Citation2014). The cp genome of Aesculus wangii (NC 035955) was included as an outgroup, and those of 32 related taxa, 30 released data from the genus Acer (Zhou et al. Citation2016; Xu et al. Citation2017; Dong et al. Citation2019; Kim et al. Citation2019; Ling & Zhang Citation2019; Shi et al. Citation2020; Liu et al. Citation2022; Wang et al. Citation2022), and both Dipteronia dyeriana (NC 031899) and D. sinensis (MK 193760) were also analyzed together (Supplementary table 1).
Figure 1. Photographs of Acer pseudosieboldianum taken by Mr. S. H. Park at the same place of sample collection site: (A) plant and (B) leaves with nine lobes. It is generally small tree up to 8 m tall and has deciduous palmate leaves with 9–11 lobes. The number of lobes are one of key characters for distinguishing the species from the related taxa in Korea. The leaves are densely pubescent when young and leaf blade is abaxially light green but adaxially dark green.

Results and discussion
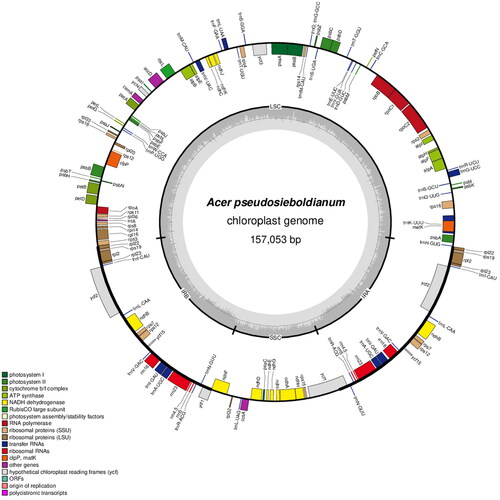
The complete chloroplast genome of A. pseudosieboldianum (NC 046487, , Figure S2) is a typical circular form composed of 157,053 bp in length with two inverted repeats (IRs) (26,747 bp) between a large single-copy (LSC) (85,391 bp) and a small single-copy (SSC) (18,168 bp) with 37.8% GC contents. It was composed of 134 genes and they identified 86 coding genes, eight rRNA genes, 37 tRNA genes, and two pseudogenes of rps2 with a 15-bp deletion in LSC and ycf1 in the boundary area of IR.
Figure 2. Complete chloroplast genome map of Acer pseudosieboldianum, containing six tracks. From the center, the first track shows the dispersed repeats which consist of direct (red) and palindromic (green) repeats. The second and third tracks show the long and short tandem repeats, respectively. The regional composition of the genome, LSC, SSC, and IRs, is identified on the fourth track. In the fifth track, the GC content along the genome is plotted. The genes are shown on the outer sixth track.
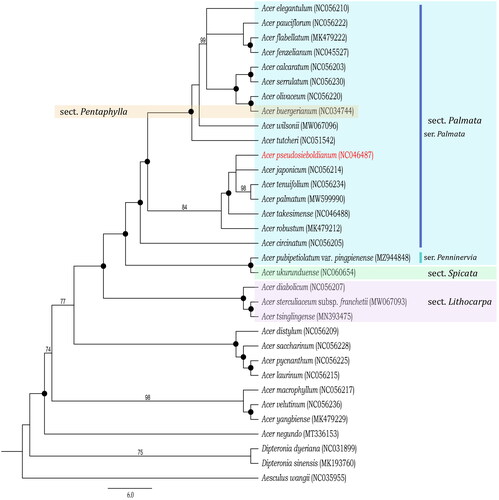
The monophyly of the genus was strongly supported in the phylogenetic tree conducted with the plastid genome sequences of 30 released data from the genus Acer and two genomes of close related genera Dipteronia, and the sister relationship between the genus Acer and Dipteronia was also cleared (). Within the genus, A. pseudosieboldianum was embedded in the series Palmata of section Palmata with its related species, though the section Palmata was paraphyletic because the series Penninervia formed a clade with section Spicata. Section Lithocarpa was sister to the sections Palmata + Spicata. In addition, A. buergerianum, a member of section Pentaphylla, was included in the section Palmata clade (). These results indicate that the infra-sectional classification within section Palmata needs to be reconsidered due to the paraphyletic relationship of A. ukurunduense (series Penninervia).
Figure 3. Phylogenetic tree of Acer pseudosieboldianum and related taxa inferred from maximum likelihood based on the 78 coding genes sequences. The outgroup was Aesculus wangii (NC 035955). The bootstrap supporting values are described on the branches, with black spots indicating 100% BP values. The position of A. pseudosieboldianum is indicated in red and the sectional classification of A. pseudosieboldianum and its related taxa are shown in different colors. The details of cp genome sequences of 30 Acer and two Dipteronia for the present molecular phylogenetic analysis are provided in Supplementary Table 1.
Conclusions
The complete chloroplast genome of the Korean maple tree Acer pseudosieboldianum is presented for the first time in this study. It has a typical circular form, composed of 157,053 bp and 134 genes. Based on the molecular phylogenetic analysis using cp genome sequence data, the phylogenetic position of A. pseudosieboldianum was confirmed within the series Palmata of section Palmata in the genus. However, the sectional classification of the genus needs to be reconsidered due to the paraphyly of section Palmata.
Author contributions
Conceiving and designing, JSK and HTK; performing and analyzing data, HTK; writing – original draft preparation, JSK and HTK; writing – review and editing, JSK; supervision, JSK; all authors have read and agreed to the published version of the manuscript.
Ethical approval
Acer pseudosieboldianum is not an endangered or protected species; therefore, specific permission was not required to collect this species. Research on this species, including the collection of plant material, was conducted following the guidelines provided by Kyungpook National University.
Supplemental Material
Download MS Word (16.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are available in GenBank of NCBI under the accession no. NC 046487 (https://www.ncbi.nlm.nih.gov/nucleotide/NC_046487.1). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA914786, SRR22857887, and SAMN32358162, respectively.
Additional information
Funding
References
- Areces-Berazain F, Wang Y, Hinsinger DD, Strijk JS. 2020. Plastome comparative genomics in maples resolves the infrageneric backbone relationships. Peer J. 8:e9483.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Dong P-B, Liu Y, Gao Q-Y, Yang T, Chen X-Y, Yang J-Y, Shang Q-H, Fang M-F. 2019. Characterization of the complete plastid genome of Acer tsinglingense, an endemic tree species in China. Mitochondrial DNA B Resour. 4(2):4065–4066.
- Grimm GW, Renner SS, Stamatakis A, Hemleben V. 2006. A nuclear ribosomal DNA phylogeny of Acer inferred with maximum likelihood, splits graphs, and motif analysis of 606 sequences. Evol Bioinform Online. 2:117693430600200.
- Kearse M, Moir R, Wilson A, Stone-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim HT, Chase MW. 2017. Independent degradation in genes of the plastid ndh gene family in species of the orchid genus Cymbidium (Orchidaceae; Epidendroideae). PLoS One. 12(11):e0187318.
- Kim HT, Pak J-H, Kim JS. 2019. The complete chloroplast genome sequence of Acer takesimense (Sapindaceae), an endemic to Ullenung Island of Korea. Mitochondrial DNA B. 4(1):1531–1532.
- Li J, Stukel M, Bussies P, Skinner K, Lemmon AR, Lemmon EM, Brown K, Bekmetjev A, Swenson NG. 2019. Maple phylogeny and biogeography inferred from phylogenomic data. J Syst Evol. 57(6):594–606.
- Li J, Yue J, Shoup S. 2006. Phylogenetics of Acer (Aceroideae, Sapindaceae) based on nucleotide sequences of two chloroplast non-coding regions. Harvard Pap Bot. 11(1):101–115.
- Ling L-Z, Zhang S-D. 2019. The complete chloroplast genome of an endangered and endemic species, Acer yangbiense (Aceraceae). Mitochondrial DNA B Resour. 5(1):224–225.
- Liu D, Chen S, Sun W. 2022. The complete chloroplast genome of Acer pubipetiolatum var. pingpienense (Sapindaceae). Mitochondrial DNA B Resour. 7(8):1448–1450.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Shi Z, Sun B, Pei N, Shi X. 2020. The complete chloroplast genome of Acer tutcheri Duthie (Acereae, Sapindaceae): an ornamental tree endemic to China. Mitochondrial DNA B Resour. 5(3):2686–2687.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Suh Y, Heo K, Park CW. 2000. Phylogenetic relationships of Maples (Acer L.; Aceraceae) implied by nuclear ribosomal ITS sequences. J Plant Res. 113(2):193–202.
- Wang X, Niu H, Huang Z. 2022. The complete chloroplast genome of Acer negundo (Aceraceae). Mitochondrial DNA B Resour. 7(1):56–57.
- Xu J-H, Wu H-B, Gao L-Z. 2017. The complete chloroplast genome sequence of the threatened trident maple Acer buergerianum (Aceraceae). Mitochondrial DNA B Resour. 2(1):273–274.
- Yu T, Gao J, Liao P-C, Li J-Q, Ma W-B. 2021. Insights into comparative analyses and phylogenomic implications of Acer (Sapindaceae) inferred from complete chloroplast genomes. Front Genet. 12:791628.
- Zhou T, Chen C, Wei Y, Chang Y, Bai G, Li Z, Kanwal N, Zhao G. 2016. Comparative transcriptome and chloroplast genome analyses of two related Dipteronia species. Front Plant Sci. 7:1512.
