Abstract
Lithocarpus konishii, a rare species endemic to islands in South China, was evaluated as a vulnerable species (VU) by the ‘China Species Red List.’ Here, we first presented the complete chloroplast genome sequence of L. konishii. The chloroplast genome was 161,059 bp in length with 36.76% GC content, containing a small single-copy region (SSC, 18,967 bp), a large single-copy region (LSC, 90,250 bp), and a pair of inverted repeats (IRs, 25,921 bp each). A total of 139 genes were predicted, including 87 protein-coding genes (CDS), 8 rRNAs, and 44 tRNAs. Based on the concatenated shared unique CDS sequence dataset, maximum-likelihood and Bayesian inference methods were used to build the phylogenetic trees of 18 species from the Fagaceae family. Results indicated that L. konishii is closely related to L. longnux and L. pachyphyllus var. fruticosus, and forms a monophyly of the subfamily Castaneoideae with Castanopsis and Castanea. This study provides a theoretical basis for the conservation genomics of this endangered plant.
Introduction
Lithocarpus konishii (Hayata) Hayata 1917 is a small evergreen tree belonging to the Fagaceae family, and is endemic to South China (Huang et al. Citation1999). It is found sporadically in a few island habitats in Hainan, Zhuhai, Hong Kong, and Taiwan, indicating a typical island disjunctive distribution (Shi et al. Citation2016). Lithocarpus konishii typically grows between 4 and 9 meters in height, with papery leaf blade, acute to caudate-acuminate apex, 3–6 obtuse teeth leaf margin, depressed globose nut, and discoid cupule (). It blossoms twice a year in April and August, with fruits ripening from July to October (Huang et al. Citation1999; Hung et al. Citation2005). L. konishii exhibits exceptional resilience to salt, alkali, drought, and sterile conditions. Additionally, it has robust wind resistance capabilities and plays a vital role in preserving water and soil within island ecosystems (Shi et al. Citation2016). Furthermore, L. konishii holds significant economic value and shows promising potential for development. For instance, the fruits are consumed by residents in Hainan after cooking, while in Taiwan, it has been cultivated as a landscaping tree and a source for truffle reproduction (You Citation2021).
Figure 1. Lithocarpus konishii (a) Natural habitat; (b) the entire plant; (c) leaf; (d) flower; (e) fruit; and (f) twig. The images were taken by Shi Shi on Dangan Island, Guangdong Province (114°14'48.89" E, 22°02'13.43" N).

However, habitat destruction and excessive deforestation have resulted in grave habitat fragmentation, population decline, and wild germplasm resources reduction of L. konishii. Based on a study utilizing chloroplast DNA atpB-rbcL for the genetic diversity analysis of L. konishii in Taiwan, it was found that the habitat of this species was severely damaged by the 1999 Chi-Chi earthquake, which resulted in a substantial loss of genetic diversity, placing L. konishii on the brink of endangerment in Taiwan. (Hung et al. Citation2005). Moreover, in the eastern part of Hainan, the population of L. konishii has been strongly affected by human activities. The expansion of roads, farmland, and housing have directly encroached upon its habitat (Shi et al. Citation2016). Due to the scarcity of wild populations of L. konishii, it has been evaluated as a vulnerable species (VU) and listed in the 'China Species Red List’ (Wang and Xie Citation2004). However, few studies have focused on the maintenance and conservation of this species. This study sequenced the complete chloroplast genome of L. konishii, and explored its phylogenetic relationships with other species in the Fagaceae family, which could be valuable to the effective utilization and protection of this species, as well as the further phylogenetic studies of this family.
Materials
Fresh and young leaves of L. konishii were collected from Dangan Island, Guangdong Province, China (114°14′48.89″ E, 22°02′13.43″ N). The voucher specimen of the sample was deposited at the Herbarium of South China Agricultural University (CANT, https://nbb.scau.edu.cn/) under the accession number 32206 (contact person: Mingxuan Zheng, [email protected]).
Methods
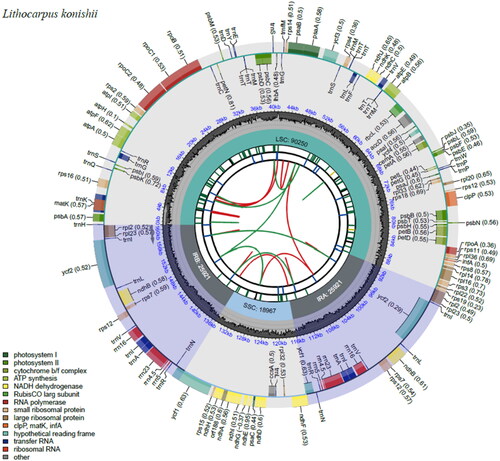
The total genomic DNA was extracted from fresh leaves using a modified cetyl trimethyl ammonium bromide (CTAB) method (Doyle and Doyle Citation1987). A genomic library consisting of an insert size of 300 bp was established by using a TruSeq DNA Sample Prep Kit (Illumina, USA) and sequenced on the Illumina Novaseq platform (Guangzhou Jierui Biotech). 5 Gb of raw data of 150 bp paired-end reads were obtained and further assembled using GetOrganelle v.1.7.7.0 (Jin et al. Citation2020). The GeSeq was used for chloroplast genome annotation (Tillich et al. Citation2017), whilst CPGAVAS2 was used to correct the annotated genome (Shi et al. Citation2019), after which it was manually checked by comparison against the complete cp genome of Lithocarpus hancei (GenBank accession number: MW375417) using Geneious v.9.0.2 (Biomatters, https://www.geneious.com) (Kearse et al. Citation2012). The complete chloroplast genome of L. konishii was submitted to GenBank with the accession number ON422319. The chloroplast Genome Viewer (CPGView, www.1kmpg.cn/cpgview/) (Liu et al. Citation2023) was used to visualize the structural features of L. konishii ().
Figure 2. Genome map of Lithocarpus konishii. The map contains six tracks by default. From the center outward, the first track shows the dispersed repeats, which consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. Black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The bottom left corner shows the functional classifications of the genes.
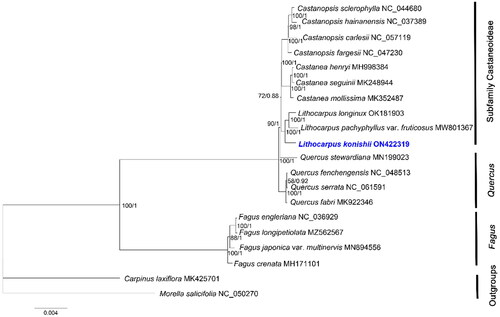
To further understand the intrageneric phylogenetic relationship of L. konishii, the complete chloroplast genome sequences of 18 species of family Fagaceae and 2 outgroup species (Morella salicifolia and Carpinus laxiflora) from the National Center for Biotechnology Information (NCBI) were aligned using MUSCLE v.3.8.31 (Edgar Citation2004). Based on the concatenated shared unique CDS sequence dataset, the phylogenetic trees were constructed using the maximum likelihood (ML) method by IQ-TREE v.2.0.3 (Nguyen et al. Citation2015) and Bayesian Inference (BI) method by Mrbayes v.3.2.6 (Ronquist et al. Citation2012). For ML analysis, a best-fit model K3Pu + F + I was selected, and the reliability of the phylogenetic tree topology was evaluated with 1,000 repeated self-expanding analyses; while for BI analysis, a best-fit model GTR + G + I was estimated by ModelTest-NG v.0.1.7 (Darriba et al. Citation2020) on the CIPRES Science Gateway (http://www.phylo.org/portal2/), the Markov Chain Monte Carlo (MCMC) was conducted for 5,000,000 generations and sampled every 100 iterations with the first 20% discarded. Branch supports were tested using the ultrafast bootstrap (UFBoot) (Hoang et al. Citation2018).
Results and discussion
The minimum and average coverage of the assembled chloroplast genome were 97× and 345.71×, respectively (Supplementary Figure 1). The complete chloroplast genome of L. konishii (ON422319) was 161,059 bp in length, presenting a typical quadripartite structure containing a small single-copy region (SSC,18,967 bp) and a large single-copy region (LSC, 90,250 bp), separated by a pair of inverted repeats (IRs, 25,921 bp each). The overall GC content of the cp genome was 36.76%. The cp genome encoded 139 genes, including 87 protein-coding genes (CDS), 8 rRNA genes, and 44 tRNA genes. There were 19 genes (rps16, atpF, rpoC1, petB, petD, rp116, rp12, ndhB, ndhA, ndhB_copy2, rp12_copy2, trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC, trnA-UGC_copy2, trnI-GAU_copy2) containing 1 intron and 4 genes (rps12, ycf3, rps12_copy2, clpP) containing 2 introns (Supplementary Figure 2), while the gene structure of the trans-splicing gene rps12 was identified, which has 3 unique exons, 2 of them are duplicated as they are located in the IR regions (Supplementary Figure 3).
Both the ML and BI trees displayed identical topologies, here we presented the ML tree (). The phylogenetic analysis shows that L. konishii is closely related to L. longnux and L. pachyphyllus var. fruticosus, and is the sister clade of Castanopsis and Castanea, forming a monophyly of the subfamily Castaneoideae, in agreement with the findings based on the nuclear and chloroplast DNA sequence data by Manos et al. (Citation2001) and the morphological data by Wang and Bo (Citation2004). Fagus represented an early branch within the family, consistent with the previous phylogeny studies of Fagaceae (Manos et al. Citation2001; Yan Citation2021; Wang and Bo Citation2004). These findings can serve as a valuable chloroplast genome resource for genetic research on germplasm resources update and afforestation tree species in the future.
Figure 3. The phylogenetic tree based on the complete chloroplast genome sequences of 18 species from the Fagaceae family, with Morella salicifolia and Carpinus laxiflora as Outgroups. The following sequences were used: Castanopsis sclerophylla NC_044680 (Ye, Hu, et al. Citation2019), Castanopsis hainanensis NC_037389 (Chen et al. Citation2018), Castanopsis carlesii NC_057119 (Sun et al. Citation2019), Castanopsis fargesii NC_047230 (Ye, Guo, et al. Citation2019), Castanea henryi MH998384 (Gao et al. Citation2019), Castanea seguinii MK248944 (Chen et al. Citation2019), Castanea mollissima MK352487 (Zhu et al. Citation2019), Lithocarpus longinux OK181903 (Wu et al. Citation2022), Lithocarpus pachyphyllus var. fruticosus MW801367 (Jin et al. Citation2021), Quercus stewardiana MN199023 (Li et al. Citation2020), Quercus fenchengensis NC_048513 (Hu et al. Citation2019), Quercus serrata NC_061591 (Yang et al. Citation2021), Quercus fabri MK922346 (Xu et al. Citation2019), Fagus engleriana NC_036929 (Yang et al. Citation2018), Fagus longipetiolata MZ562567 (Liang et al. Citation2022), Fagus japonica var. multinervis MN894556 (Park and Oh Citation2020), Fagus crenata MH171101 (Worth et al. Citation2019), Carpinus laxiflora MK425701 (Lee et al. Citation2019), and Morella salicifolia NC_050270 (Karumuna et al. Citation2019). NCBI accession numbers of each genome are shown in the figure. The bootstrap support values were based on 1000 replicates. Values of bootstrap support and posterior probability for each branch nodes are as indicated.
Ethical approval
This study was ethically approved and received permission for the sample collection from the Guangdong Zhuhai Qi’ao-Dangan Island Provincial Level Nature Reserve (Admission number: 07103300202302110900180053439). The fieldwork of this study was supported and assisted by the staff from Zhuhai Qi’ao-Dangan Island Provincial Level Nature Reserve. Lithocarpus konishii is vulnerable (VU) according to China species red list: Vol. I red list (2004), therefore all the collection and experimental work complied with policies research involving species at risk of extinction described in ‘the Regulations of the People’s Republic of China on Wild Plant Protection’ and ‘the International Union for Conservation of Nature.’
Author contributions
Shi Shi and Mingyan Ding designed the study. Mingyan Ding, Hongkang Shen, Zhilai Yang and Shi Shi collected the materials. Keyi Fu, Mingyan Ding, Zhilai Yang and Linjin Lu performed the experiments and analysis. Keyi Fu and Hongkang Shen drafted the original manuscript. Shi Shi, Zhilai Yang and Mingyan Ding revised the manuscript. All authors approved the final version of the manuscript and agreed to be accountable for all aspects of the work.
Supplemental Material
Download JPEG Image (172.7 KB)Supplemental Material
Download PNG Image (531.7 KB)Supplemental Material
Download TIFF Image (1.8 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at [https://www.ncbi.nlm.nih.gov/], reference number ON422319. The associated “BioProject”, “Bio-Sample” and “SRA” numbers are PRJNA838448, SAMN28422863 and SRR19233775 respectively.
Additional information
Funding
References
- Chen X, Liu QZ, Guo W, Xu L, Zhang LS. 2019. The complete chloroplast genome of Castanea seguinii, an endemic to China. Mitochondrial DNA Part B. 4(1):758–759. doi: 10.1080/23802359.2019.1565970.
- Chen XD, Yang J, Yang YC, Zhang X, Du XM, Zhao GF. 2018. Characterization of the complete plastid genome of Castanopsis hainanensis Merrill. Conservation Genet Resour. 10(4):825–828. doi: 10.1007/s12686-017-0940-9.
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294. doi: 10.1093/molbev/msz189.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Edgar RC. 2004. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. doi: 10.1093/nar/gkh340.
- Gao XX, Yan F, Liu M, Zulfiqar S, Zhao P. 2019. The complete chloroplast genome sequence of an endemic species Pearl chestnut (Castanea henryi). Mitochondrial DNA Part B. 4(1):551–552. doi: 10.1080/23802359.2018.1553522.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi: 10.1093/molbev/msx281.
- Hu HL, Wang LZ, Yang J, Zhang RS, Li Q, Liu YQ, Qin L. 2019. The complete chloroplast genome of Quercus fenchengensis and the phylogenetic implication. Mitochondrial DNA B Resour. 4(2):3066–3067. doi: 10.1080/23802359.2019.1666040.
- Huang CJ, Zhang YT, Bartholo B. 1999. Flora of China. Vol. 4. Science Press; p. 358.
- Hung KH, Hsu TW, Schaal BA, Chiang TY. 2005. Loss of genetic diversity and erroneous phylogeographical inferences in Lithocarpus konishii (Fagaceae) of Taiwan caused by the Chi-Chi earthquake: implications for conservation. Ann Missouri Botan Garden. 92(1):52–65. DOI: http://www.jstor.org/stable/3298648.
- Jin JJ, Yu WB, Yang JB, Song Y, De Pamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31. doi: 10.1186/s13059-020-02154-5.
- Jin L, Liu JJ, Xiao TW, Li QM, Lin LX, Shao XN, Ma CX, Li BH, Mi XC, Qiao XJ, et al. 2021. Community phylogenetics require phylogenies reconstructed from plastid genomes. Authorea Preprints. doi: 10.22541/au.161834751.14170237/v1.
- Karumuna JJ, Yan DY, Kyalo CM, Li ZZ. 2019. The complete chloroplast genome sequence of Morella salicifolia (Myricaceae): characterization and phylogenetic analysis. Mitochondrial DNA Part B. 4(1):963–964. doi: 10.1080/23802359.2019.1580157.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Lee MW, Kim SC, Ahn JY, Lee JW. 2019. The complete chloroplast genome of Carpinus Laxiflora (Betulaceae). Mitochondrial DNA Part B. 4(1):1643–1644. doi: 10.1080/23802359.2019.1604184.
- Li Y, Wang L, Zhao Y, Fang YM. 2020. The complete chloroplast genome sequence of Quercus stewardiana (Fagaceae). Mitochondrial DNA Part B. 5(2):1958–1959. doi: 10.1080/23802359.2020.1756961.
- Liang DQ, Wang HY, Zhang J, Zhao YX, Wu F. 2022. Complete chloroplast genome sequence of Fagus longipetiolata Seemen (Fagaceae): Genome structure, adaptive evolution, and phylogenetic relationships. Life. 12(1):92. doi: 10.3390/life12010092.
- Liu SY, Ni Y, Li JL, Zhang XY, Yang HY, Chen HM, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Manos PS, Zhou ZK, Cannon CH. 2001. Systematics of Fagaceae: phylogenetic tests of reproductive trait evolution. Int J Plant Sci. 162(6):1361–1379. doi: 10.1086/322949.
- Nguyen LT, Schmidt HA, Arndt VH, Quang MB. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Park JS, Oh SH. 2020. A second complete chloroplast genome sequence of Fagus multinervis Nakai (Fagaceae): intraspecific variations on chloroplast genome. Mitochondrial DNA Part B. 5(2):1868–1869. doi: 10.1080/23802359.2020.1752837.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. System Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345.
- Shi S, Fan Q, Chen SF, Tan WZ. 2016. Study on the biogeographic pattern of the discontinuous distribution between Hainan and Taiwan – based on the genetic geography analysis of Coleoptera. Beijing: China Science and Technology (paper online)
- Sun RX, Ye XM, Wang ZL, Lin XF. 2019. The complete chloroplast genome of Castanopsis carlesii (Hemsl.) Hay. Mitochondrial DNA B Resour. 4(2):2591–2592. doi: 10.1080/23802359.2019.1641437.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Wang PL, Bo FD. 2004. Pollen morphology and biogeography of Fagaceae plants. Guangzhou: Guangdong Science and Technology Press. p. 7–107.
- Wang S, Xie Y. 2004. China species red list: vol. I red list. Beijing: Higher Education Press. p. 314–315.
- Worth JRP, Liu L, Wei FJ, Tomaru N. 2019. The complete chloroplast genome of Fagus crenata (subgenus Fagus) and comparison with F.engleriana (subgenus Engleriana). PeerJ. 7:e7026. doi: 10.7717/peerj.7026.
- Wu CY, Lin L, Yao KP, Yang RJ, Deng M. 2022. The complete chloroplast genome sequence of Lithocarpus longinux (Fagaceae). Mitochondrial DNA B Resour. 7(7):1229–1231. doi: 10.1080/23802359.2022.2093664.
- Xu Y, Chen H, Qi M, Su W, Zhang Y, Du FK. 2019. The complete chloroplast genome of Quercus fabri (Fagaceae) from China. Mitochondrial DNA B Resour. 4(2):2857–2858. doi: 10.1080/23802359.2019.1660921.
- Yan ZT. 2021. Research progress of Fagus plants in China. Agric Develop Equip. 240(12):107–108.
- Yang YC, Zhou T, Qian ZQ, Zhao GF. 2021. Phylogenetic relationships in Chinese oaks (Fagaceae, Quercus): Evidence from plastid genome using low-coverage whole genome sequencing. Genomics. 113(3):1438–1447. doi: 10.1016/j.ygeno.2021.03.013.
- Yang YC, Zhu J, Feng L, Zhou T, Bai GQ, Yang J, Zhao GF. 2018. Plastid genome comparative and phylogenetic analyses of the key genera in Fagaceae: highlighting the effect of codon composition bias in phylogenetic inference. Front Plant Sci. 9:82. doi: 10.3389/fpls.2018.00082.
- Ye XM, Guo YP, Lei XG, Sun RX. 2019a. The complete chloroplast genome of Castanopsis Fargesii Franch. (Fagaceae). Mitochondrial DNA Part B. 4(1):1656–1657. doi: 10.1080/23802359.2019.1605850.
- Ye XM, Hu DG, Guo YP, Sun RX. 2019b. Complete chloroplast genome of Castanopsis sclerophylla (Lindl.) Schott: genome structure and comparative and phylogenetic analysis. PLOS One. 14(7):e0212325. doi: 10.1371/journal.pone.0212325.
- You SF. 2021. The new species Lithocarpus konishii truffle published by Taiwan Forestry Research Institute. Agriculture Media [accessed 2021 Mar 30]. https://www.agriharvest.tw/archives/57060.
- Zhu CC, Shi FH, Wang M, Zhao YQ, Chen Y, Geng GM. 2019. The complete chloroplast genome of a variety of Castanea mollissima 'Hongli’ (Fagaceae). Mitochondrial DNA Part B. 4(1):993–994. doi: 10.1080/23802359.2019.1580160.
