Abstract
The freshwater sleeper, Sineleotris saccharae Herre, Citation1940 is a member of the Odontobutiae family, widely distributed in southern China. In the present study, we determined the complete mitochondrial genome of S. saccharae for the first time and analyzed its evolutionary relationship. The complete mitochondrial genome of S. saccharae was 16,487 bp long, and had 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNA (rRNAs) and a control region (CR). The mitogenome of S. saccharae shared the same gene organization and orientation as other teleosts. According to phylogenetic research, S. saccharae was sister to S. chalmersi with high support value, providing the monophyly of the genus Sineleotris. These results will be helpful for understanding the systematics of the odontobutids.
Introduction
Sineleotris saccharae originally described by Herre (Citation1940), was native to Southern China (). Based on morphological traits, the species was subsequently assigned to the genera Philypnus (Chen and Zheng Citation1985), Hypseleotris (Wu Citation1991), and Sineleotris (Wu and Zhong Citation2008). Chen et al. (Citation2002) initially classified the genus Sineleotris as belonging to the Odontobutidae family, while Li et al. (Citation2018) verified this classification using molecular data. Information about genetic characteristics of S. saccharae, including mitochondrial genome, genetic diversity, is still not available. In the present study, we determined mitogenome of S. saccharae for the first time and established the phylogenetic relationship of the odontobutids.
Materials
Adult S. saccharae individuals were collected from Fengshun, Guangdong Province, China (23.817546°N, 116.287902E). A specimen and its DNA were deposited at the ichthyoligocal museum of Freshwater Fisheries Research Institute of Jiangsu Province, China (Dr Liqiang Zhong, e-mail: [email protected]) under the voucher number JSFFRI-20008.
Methods
Total DNA was extracted with Qiagen Blood and Cell Midi Kit. The mitogenome was amplified using 20 pairs of Odontobutis-specific primers (Ma et al. Citation2015) and 30 sets of fish-universal primers (Miya and Nishida Citation1999). The gaps were filled with self-designed primers (supplemental Table S1). Using the same PCR primers, the PCR products were sequenced via an Applied Biosystems ABI 3730XL capillary sequencer.
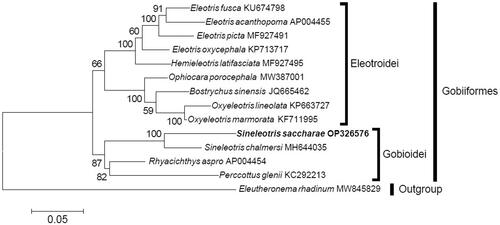
After blasting in the GenBank, raw sequencing data were assembled to final mitogenome with manually inspecting. Then MitoFish was used to annotate and visualize it (Iwasaki et al. Citation2013). To analyze the phylogenetic position of S. saccharae, 13 PCGs from the closest thirteen fishes were downloaded according to blasting results in GenBank (, 12 species of gobiiformes and an outgroup Eleutheronema rhadinum). Each of 13 PCGs was aligned separately using Clustal W with default settings and then concatenated to a single multiple sequence alignment. The substitution model mtREV + G + I were selected as the best model for analysis. The maximum likelihood (ML) analysis (Felsenstein Citation1985) was inferred on MEGA 11 (Tamura et al. Citation2021) with 1000 bootstrap replicates.
Table 1. Species and GenBank accession number of mitogenomes used in this study.
Results
The entire mitogenome of S. saccharae was 16,487 bp long, and had 13 PCGs, 22 tRNAs, 2 rRNAs and a CR (). The mitogenome of S. saccharae shared the same gene organization and orientation as other teleosts (). The overall base composition was T 25.3%, C 30.0%, A 28.9%, and G 15.8%. Only one of the 13 PCGs, the ND6, was found to be encoded on the light strand (L-strand), with the other 12 being identified on the heavy strand (H-strand). The 850-bp-long CR has the highest A + T concentration (65.0%) in the entire mitogenome.
Figure 2. Gene map of the mitochondrial genome of Sineleotris saccharae (GenBank accession number: OP326576), with 13 protein coding genes, 22 tRNAs, 2 rRNAs, and a control region. Genes encoded on light strand and heavy-strand were shown inner and outside of the ring respectively.
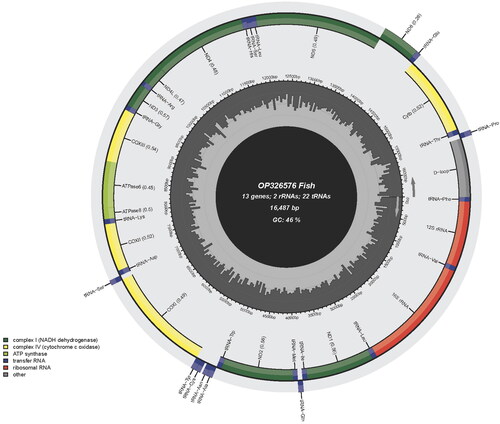
Table 2. Organization of the mitogenome of Sineleotris saccharae.
In the ML phylogenetic tree (), S. saccharae was firstly clustered with S. chalmersi with high support value, then together with Perccottus glenii and Rhyacichthys aspro forming the Gobioidei suorder (bootstrap value >80). While the remaining nine sleepers clustered together forming the Eleotroidei suorder.
Discussion and conclusion
In this study, the entire mitogenome of S. saccharae was identified for the first time. It was similar to that of other teleosts in terms of gene organization, and composition (Miya et al. Citation2003). In the ML phylogenetic analysis, S. saccharae was sister to S. chalmersi with high support value, providing the monophyly of the genus Sineleotris. And all sleepers were placed into two well-supported suborder clusters, which was similar to those of previous studies (Zhong et al. Citation2018a, Citation2018b). These results will be essential to the species identification and systematics of the odontobutids in the future.
Author contributions
ZLQ conceived this study; ZLY, LYS and CXH conducted the experiments, LDM and TSK analyzed the data; ZLY and WMH wrote the drafting of the paper; ZLQ revised it critically, and that all authors agree to be accountable for all aspects of the work.
Ethical approval
This study was approved by the animal care and Ethical Committee of Freshwater Fisheries Research Institute of Jiangsu Province.
Supplemental Material
Download MS Word (21.7 KB)Acknowledgments
We are grateful to Shujie Liu, Binbin Zhan and Huiwen Xiao for help in field assistance and the species reference image.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitochondrial genome sequence is available on GenBank of NCBI at www.ncbi.nlm.nih.gov with the accession number of OP326576.
Additional information
Funding
References
- Alda F, Adams AJ, McMillan WO, Chakrabarty P. 2017. Complete mitochondrial genomes of three Neotropical sleeper gobies: Eleotris amblyopsis, E. picta and Hemieleotris latifasciata (Gobiiformes: Eleotridae). Mitochondrial DNA B Resour. 2(2):747–750. doi: 10.1080/23802359.2017.1390412.
- Amin MH, Lee SR, Irawan B, Andriyono S, Kim HW. 2021. Characterization of the complete mitochondrial genome of the Northern Mud Gudgeon, Ophiocara porocephala (Perciformes: Eleotridae) with phylogenetic implications. Mitochondrial DNA B Resour. 6(3):953–955. doi: 10.1080/23802359.2021.1889415.
- Chen IS, Kottelat M, Wu HL. 2002. A new genus of freshwater sleeper (Teleostei: Odontobutididae) from southern China and mainland Southeast Asia. J Fish Soc Taiwan. 29:229–235.
- Chen W, Zheng CY. 1985. Three species of Eleotridae from China. J Sci Med Jinan Univ. 1:73–80.
- Felsenstein J. 1985. Confidence-limits on phylogenies-an approach using the bootstrap. Evolution. 39(4):783–791. doi: 10.2307/2408678.
- Herre AW. 1940. Notes on fishes in the Zoological Museum of Stanford University, VIII. A new genus and two new species of Chinese gobies with remarks on some other species. Philipp J Sci. 73:293–298.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540. doi: 10.1093/molbev/mst141.
- Li HJ, He Y, Jiang JM, Liu ZZ, Li CH. 2018. Molecular systematics and phylogenetic analysis of the Asian endemic freshwater sleepers (Gobiiformes: Odontobutidae). Mol Phylogenet Evol. 121:1–11. doi: 10.1016/j.ympev.2017.12.026.
- Ma ZH, Yang XF, Bercsenyi M, Wu JJ, Yu YY, Wei KJ, Fan QX, Yang RB. 2015. Comparative mitogenomics of the genus Odontobutis (Perciformes: Gobioidei: Odontobutidae) revealed conserved gene rearrangement and high sequence variations. Int J Mol Sci. 16(10):25031–25049. doi: 10.3390/ijms161025031.
- Miya M, Nishida M. 1999. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotechnol (NY). 1(5):416–0426. doi: 10.1007/pl00011798.
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26(1):121–138. doi: 10.1016/s1055-7903(02)00332-9.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi: 10.1093/molbev/msab120. 33892491
- Wang CG, Zhang M, Cheng GP, Chen XL. 2019. The complete mitochondrial genome of Microdous chalmersi (Gobiiformes: Odontobutidae). Mitochondrial DNA B. 4(1):1979–1980. doi: 10.1080/23802359.2019.1617075.
- Wu HL. 1991. The freshwater fishes of Guangdong Province. Guangzhou: Guangdong Science and Technology Press.
- Wu HL, Zhong JS. 2008. Fauna sinica, Osteichthyes, Perciformes (V), Gobioidei. Beijing: Science Press.
- Xia AJ, Zhong LQ, Chen XH, Bian WJ, Zhang TQ, Shi YB. 2015. Complete mitochondrial genome of spined sleeper Eleotris oxycephala (Perciformes, Eleotridae) and phylogenetic consideration. Biochem Syst Ecol. 62:11–19. doi: 10.1016/j.bse.2015.07.030.
- Xue W, Hou GY, Li CY, Kong XF, Zheng XH, Li JT, Sun XW. 2013. Complete mitochondrial genome of Chinese sleeper, Perccottus glenii. Mitochondr DNA. 24(4):339–341. doi: 10.3109/19401736.2012.760081.
- Yang ZY, Liang HW, Li Z, Wang D, Zou GW. 2016. Mitochondrial genome of the Marbled goby (Oxyeleotris marmorata). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1073–1074. doi: 10.3109/19401736.2014.928873.
- Zang X, Yin DQ, Wang RR, Yin SW, Tao PF, Chen JW, Zhang GS. 2016. Complete mitochondrial DNA sequence and phylogenic analysis of Oxyeleotris lineolatus (Perciformes, Eleotridae). Mitochondrial DNA A. 27(4):2414–2416. doi: 10.3109/19401736.2015.1030621.
- Zhong LQ, Wang MH, Li DM, Tang SK, Zhang TQ, Bian WJ, Chen XH. 2018a. Complete mitochondrial genome of Odontobutis haifengensis (Perciformes, Odontobutiae): A unique rearrangement of tRNAs and additional noncoding regions identified in the genus Odontobutis. Genomics. 110(6):382–388. doi: 10.1016/j.ygeno.2017.12.008.
- Zhong LQ, Wang MH, Li DM, Tang SK, Zhang TQ, Bian WJ, Chen XH. 2018b. Complete mitochondrial genome of freshwater goby Rhinogobius cliffordpopei (Perciformes, Gobiidae): genome characterization and phylogenetic analysis. Genes Genomics. 40(11):1137–1148. doi: 10.1007/s13258-018-0669-1.
- Zhong LQ, Wang MH, Li DM, Tang SK, Chen XH. 2021. Mitochondrial genome of Eleutheronema rhadinum with an additional non-coding region and novel insights into the phylogenetics. Front Mar Sci. 8:746598. doi: 10.3389/fmars.2021.746598.