Abstract
Ziziphus jujuba Mill., commonly referred to as jujube, is a species of fruiting buckthorn (family Rhamnaceae) that is frequently found across the Shaanxi, Shanxi, and Hebei provinces of China. The ‘Fengmiguan’ variety of jujube, also known as ‘Honey jar,’ is distinguished by its high yield and sugar content, as well as its strong ability to adapt to different environments. In this study, we sequenced and assembled the chloroplast genome (i.e. the plastome) of ‘Fengmiguan’ jujube using a paired-end short-read sequencing technique. The plastome exhibits a quadripartite structure with a total length of 161,818 bp that consists of a large single-copy region (89,427 bp), a small single-copy region (19,361 bp), and two inverted repeats (26,515 bp). The GC content of the plastome is 36.75%. Annotation of the ‘Fengmiguan’ jujube plastome revealed 123 genes, including 79 protein-coding genes, 36 transfer RNA genes, and eight ribosomal RNA genes. Phylogenetic analysis revealed that the ‘Fengmiguan’ variety is closely related to the ‘Bokjo’ variety. Furthermore, we found four variations between these two varieties of jujube, one of which was a 101 bp insertion. Our findings enhance the current understanding of the phylogenetic relationship between different varieties of Z. jujuba Mill., which could possibly aid in the improvement of genetic breeding and population selection in jujubes.
Introduction
Jujube (Ziziphus jujuba Mill.) is a species of fruiting buckthorn (family Rhamnaceae) that grows well in cold, temperate, and warm climates, and is one of the most important species of the Rhamnaceae family due to its economic, ecological, and social importance (Liu et al. Citation2020). Jujube has a long history of cultivation in China and is a popular food product due to its high levels of nutrients, including vitamin C and organic acids (Shan et al. Citation2019). The ‘Fengmiguan’ variety of jujube, also known as ‘Honey jar,’ is characterized by its small fruit and high sugar content and is thus ideal for fresh consumption or dehydration (Yao and Guldan Citation2012). In recent years, the chloroplast genomes of several jujube varieties have been sequenced and subjected to phylogenetic analyses (Zhang et al. Citation2021). However, the chloroplast genome of the ‘Fengmiguan’ variety has not yet been reported; therefore, it is necessary to decode the chloroplast genome of this variety and compare it to that of other jujube varieties or related species. In this study, we sequenced the chloroplast genome of ‘Fengmiguan’ jujube for the first time and determined its phylogenetic position within the Rhamnaceae family; this will provide important information about the ‘Fengmiguan’ jujube plastome that will be valuable for genetic breeding and population selection.
Materials
We collected 30 g of fresh ‘Fengmiguan’ jujube leaves from the experimental base of Hebei Agriculture University, Baoding, Hebei Province, China (115.425°E, 38.815°N, Alt. 79.8 m) (). The leaf samples are preserved at the Research Center of Chinese Jujube, Hebei Agriculture University (https://zgzyjzx.hebau.edu.cn/zxgk/zxjj.htm, contact Lu Han, [email protected]), and DNA samples were stored under the ID number JJB150.
Methods
DNA isolation
A modified cetyltrimethylammonium bromide method (Clarke Citation2009) was used to extract whole genomic DNA from the leaf samples. DNA purity was assessed using a NanoDrop One UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and DNA integrity was verified by agarose gel electrophoresis.
Library preparation, sequencing, and annotation
The isolated DNA was used for paired-end library construction with an insert size of 200–400 bp, using the official standard protocol, and sequenced on the MGISEQ-2000 platform (BGI, Shenzhen, Guangdong, China). The clean paired-end reads were then assembled using GetOrganelle v1.7.6.1 software (Jin et al. Citation2020) with the Z. jujuba (KU351660) genome serving as a reference. We annotated the resulting chloroplast genome using CPGAVAS2 software (Shi et al. Citation2019) and manually corrected it using Geneious Prime software (v2021.1.1).
Phylogenetic analysis
We aligned 31 complete chloroplast sequences using MAFFT (v7.505) (Katoh and Standley Citation2013) and constructed a maximum-likelihood phylogenetic tree (1000 bootstrap replications; JTT model) using MEGA software (v11.0.13) (Tamura et al. Citation2007).
Screening of variant sites
We used msa2vcf software (https://github.com/connor-lab/msa2vcf) to differentiate between single nucleotide polymorphisms and insertion–deletions.
Results
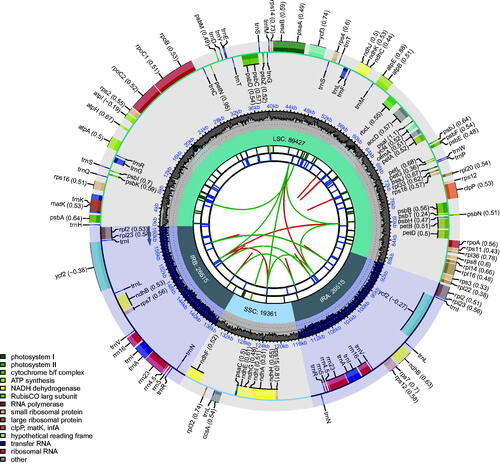
The chloroplast genome of ‘Fengmiguan’ jujube is made up of 161,818 bp and exhibits a quadripartite structure, consisting of a large single-copy (LSC) region (89,427 bp), a small single-copy (SSC) region (19,361 bp), and two inverted repeats (26,515 bp each) (). The GC content of the ‘Fengmiguan’ jujube plastome is 36.75% (). We detected 123 chloroplast genes, including 79 protein-coding genes, 36 transfer RNA genes, and eight ribosomal RNA genes. Among the annotated genes, only one (rps12) exhibited trans-splicing, and 10 (ycf3, clpP, petB, rps16, rpoC1, ndhB, ndhA, petD, rpl16, and rpl2) exhibited cis-splicing.
Figure 2. Schematic of the ‘Fengmiguan’ jujube chloroplast genome. The map contains six tracks. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, type of repeat they represent, and the description of the repeat types are as follows: black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. Genes are color-coded according to their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The key for the functional classification of the genes is shown in the bottom left corner.
To determine the phylogenetic position of ‘Fengmiguan’ jujube within the Rhamnaceae family, we performed phylogenetic analysis of the chloroplast genomes from two outgroups (Elaeagnus conferta and Elaeagnus umbellata) and one Berchemiella, two Ventilago, four Rhamnus, three Berchemia, four Hovenia, four Spyridium, and 11 Ziziphus species (; ). Phylogenetic analyses revealed that the ‘Fengmiguan’ variety is closely related to the ‘Bokjo’ variety and distantly related to the Ziziphus mauritiana and Ziziphus spina-christi varieties. Upon comparing chloroplast genomes of the ‘Fengmiguan’ and ‘Bokjo’ genotypes, we discovered four variations within the intergenic region of the LSC region, including two deletions (‘T’ and ‘A’ at positions 34,560 and 54,160, respectively) and two insertions (‘ATTGT’ at position 60,510 and a 101 bp insertion at the 61,067 position). The 101 bp insertion is the primary reason for the difference in length between this ‘Fengmiguan’ genotype and the published ‘Bokjo’ genotype, with lengths of 161,818 bp and 161,714 bp, respectively. Furthermore, we compared the complete chloroplast genome sequences of all five representative jujube genotypes available in the National Center for Biotechnology Information database. Our analysis revealed that this ‘Fengmiguan’ genotype differs from the other jujube chloroplast-available genotypes due to the presence of the 101 bp insertion.
Figure 3. The phylogenetic tree including two outgroups (Elaeagnus conferta and Elaeagnus umbellata) and one Berchemiella, two Ventilago, four Rhamnus, three Berchemia, four Hovenia, four Spyridium, and 11 Ziziphus species. This tree is based on DNA sequences and was constructed using the maximum-likelihood (ML) method. The numbers next to each node indicate the percentage of bootstrap support for 1000 replicates, and all nodes without numbers are 100% supported by the bootstrap sampling test.
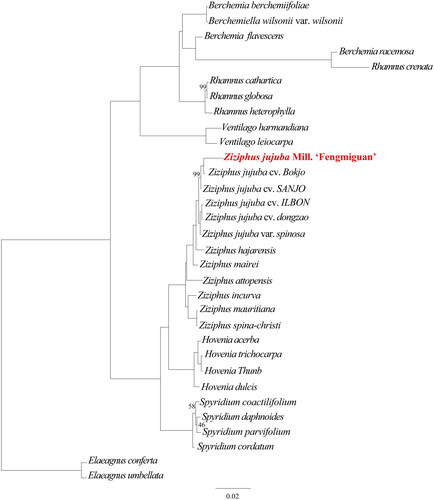
Table 1. NCBI accession numbers and citations for the sources of the 31 species used in the construction of the phylogenetic tree.
Discussion and conclusions
In this study, we sequenced, assembled, and annotated the chloroplast genome of one genotype from the Z. jujuba Mill. ‘Fengmiguan’ variety. This variety of jujube is famous for its sweet flavor and crisp texture, as well as its abundant fruit production. The chloroplast genome is a suitable model for phylogenetic investigations, due to its specificity (Hu et al. Citation2022). The comparison of ‘Fengmiguan’ and ‘Bokjo’ plastomes revealed four variations that were present in the ‘Fengmiguan’ plastome, including two deletions and two insertions. These findings contribute to our understanding of the phylogenetic relationship between different Z. jujuba Mill. varieties, which will facilitate improved genetic breeding and population selection in jujubes.
Author contributions
MY and ML conceived the study. SZ and BL collected the samples and performed the experiments. MY, SZ, BL, and LH analyzed the data and prepared the manuscript. MY and ML revised the manuscript. All authors have reviewed and approved the manuscript.
Ethical approval
Research on the chloroplast genome of plants does not require ethical permission or approval.
Disclosure statement
No potential conflict of interest was reported by any of the authors.
Data availability statement
The genome sequence data obtained in this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number OP537226. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA884017, SAMN31006321, and SRR22243258, respectively.
Additional information
Funding
References
- Asaf S, Ahmad W, Al-Harrasi A, Khan AL. 2022. Uncovering the first complete plastome genomics, comparative analyses, and phylogenetic dispositions of endemic medicinal plant Ziziphus hajarensis (Rhamnaceae). BMC Genomics. 23(1):83. doi: 10.1186/s12864-022-08320-2.
- Cheon KS, Kim KA, Yoo KO. 2018. The complete chloroplast genome sequence of Berchemia berchemiifolia (Rhamnaceae). Mitochondrial DNA B Resour. 3(1):133–134. doi: 10.1080/23802359.2018.1431068.
- Clarke JD. 2009. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb Protoc. 2009(3). doi: 10.1101/pdb.prot5177.
- Clowes C, Fowler RM, Brown GK, Bayly MJ. 2018. The complete chloroplast genome sequence of Spyridium parvifolium var. parvifolium (family Rhamnaceae; tribe Pomaderreae). Mitochondrial DNA B Resour. 3(2):807–809. doi: 10.1080/23802359.2018.1483776.
- Gao CM, Gao YC, Liu XH. 2017. The complete genome of Ziziphus jujuba cv. dongzao, an economic crop in Yellow River Delta of China. Mitochondrial DNA B Resour. 2(2):692–693. doi: 10.1080/23802359.2017.1383206.
- Hu G, Wu Y, Guo C, Lu D, Dong N, Chen B, Qiao Y, Zhang Y, Pan Q. 2022. Haplotype analysis of chloroplast genomes for jujube breeding. Front Plant Sci. 13:841767. doi: 10.3389/fpls.2022.841767.
- Huang J, Chen R, Li X. 2017. Comparative analysis of the complete chloroplast genome of four known Ziziphus species. Genes. 8(12):340. doi: 10.3390/genes8120340.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31. doi: 10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kim M, Park JH, Gil J, Kim JS, Kim HT, Oh HK, Lee KH, Lee M, Lee J, Lee Y, et al. 2022. The complete chloroplast genome of Ziziphus jujuba cv. Bokjo (Rhamnaceae). Mitochondrial DNA Part B. 7(10):1805–1806. doi: 10.1080/23802359.2022.2131366.
- Li J. 2021. The complete chloroplast genome sequence of Ziziphus attopensis (Rhamnaceae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 6(10):2828–2829. doi: 10.1080/23802359.2021.1915716.
- Li M, Ye X, Bi H. 2020. Characterization of the complete chloroplast genome of two Hovenia species (Rhamnaceae). Mitochondrial DNA Part B. 5(2):1731–1732. doi: 10.1080/23802359.2020.1749177.
- Liu D, Tong B-Q, Li W-Q, Wang L, Xian Y, Han B, Dong X, Lu Y-Z, Li W, Xie X-M, et al. 2021. The first complete chloroplast genome of Hovenia dulcis Thunb. (Rhamnaceae). Mitochondrial DNA B Resour. 6(3):916–917. doi: 10.1080/23802359.2021.1887772.
- Liu J, Gong L-D, Qi L, Liu Z-Y, Niu Y-F, Shi C. 2019. The complete chloroplast genome of Elaeagnus conferta Roxb (Elaeagnaceae). Mitochondrial DNA Part B. 4(1):2035–2036. doi: 10.1080/23802359.2019.1617074.
- Lu X, Luo Q, Qin Y, Yan Q, Guo S. 2021. The complete chloroplast genome sequence of Ventilago leiocarpa Benth. Mitochondrial DNA B Resour. 6(3):736–737. doi: 10.1080/23802359.2020.1861559.
- Lu Y, Ma Q, Xu X, Wang Z, Andrie I, Savitskaya T, Yuzuak S. 2022. Characterization of the complete chloroplast genome of Elaeagnus pungens (Elaeagnaceae) and phylogeny within Elaeagnaceae. Mitochondrial DNA B Resour. 7(7):1213–1215. doi: 10.1080/23802359.2022.2090291.
- Liu M, Wang J, Wang L, Liu P, Zhao J, Zhao Z, Yao S, Stănică F, Liu Z, Wang L, et al. 2020. The historical and current research progress on jujube – a superfruit for the future. Hortic Res. 7:119. doi: 10.1038/s41438-020-00346-5.
- Park JM, Koo J. 2023. The complete chloroplast genome sequence of Berchemia racemosa Siebold & Zucc. (Rhamnaceae), a rare plant species in Korea. Mitochondrial DNA B Resour. 8(1):69–72. doi: 10.1080/23802359.2022.2161329.
- Shan S, Xie Y, Zhao H, Niu J, Zhang S, Zhang X, Li Z. 2019. Bound polyphenol extracted from jujube pulp triggers mitochondria-mediated apoptosis and cell cycle arrest of HepG2 cell in vitro and in vivo. J Funct Foods. 53:187–196. doi: 10.1016/j.jff.2018.12.017.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24(8):1596–1599. doi: 10.1093/molbev/msm092.
- Wang J, Yang S. 2021. The complete chloroplast genome of Rhamnus crenata Siebold & Zuccarini (Rhamnaceae). Mitochondrial DNA B Resour. 6(9):2489–2490. doi: 10.1080/23802359.2021.1945502.
- Wang YH, Chen SY, Zhang SD. 2018. Characterization of the complete chloroplast genome of Berchemiella wilsonii var. wilsonii (Rhamnaceae), an endangered species endemic to China. Conserv Genet Resour. 10(1):39–41. doi: 10.1007/s12686-017-0760-y.
- Wang Y, Hao J, Yuan X, Lu B. 2019. The complete chloroplast genome sequence of Ziziphus incurva. Mitochondrial DNA B Resour. 4(2):3465–3466. doi: 10.1080/23802359.2019.1674706.
- Xie Y, Wang Z, Jiang X, Zhang X. 2020. The complete chloroplast genome of Rhamnus globosa (Rhamnaceae). Mitochondrial DNA B Resour. 5(3):2830–2831. doi: 10.1080/23802359.2020.1791010.
- Yao S, Guldan S. 2012. Jujube—Chinese Date, a potential fruit crop in New Mexico. ASHS Conference, Alexandria.
- Zhang L, Mao R, Bi H, Shen J, Wang Y, Li M. 2020. Characterization of the complete chloroplast genome of Hovenia acerba (Rhamnaceae). Mitochondrial DNA B Resour. 5(1):934–935. doi: 10.1080/23802359.2020.1714492.
- Zhang Y, Hu G, Mao W, Dong N, Chen B, Pan Q. 2021. Chloroplast genome sequence of the wild Ziziphus jujuba Mill. var. spinosa from North China. Mitochondrial DNA B Resour. 6(2):666–667. doi: 10.1080/23802359.2021.1878962.
- Zhu XF, Li Y, Lu Z. 2019. The complete chloroplast genome sequence of Berchemia flavescens (Rhamnaceae). Mitochondrial DNA Part B. 4(1):1302–1303. doi: 10.1080/23802359.2019.1591240.

