Abstract
Myristica argentea Warb. 1891 and M. fatua Houtt. 1774 are two South-East Asian food tree species. They are harvested from the wild or cultivated for local uses as a condiment (nutmeg and mace), medicine, and source of wood. In this study, we reconstructed the complete chloroplast (cp) genomes of these two species from whole genome sequencing data using the Illumina NovaSeq platform. The genome sizes of M. argentea and M. fatua were respectively 155,871 base pairs (bp) and 155,898 bp, including 126 genes and an overall GC content of 39.20% in both species. Our study provides useful resources for future evolutionary research and diversity analysis of Myristica species.
Introduction
The Myristica genus contains 172 species that are mainly distributed in Tropical Southeast Asia, the Malay Archipelago (Malesia) and the western Pacific (de Wilde Citation2014; Govaerts et al. Citation2021). Myristica argentea Warb. 1891, known as Papuan nutmeg or Fakfak nutmeg, is endemic to Papua island, Indonesia, and is named after the region where the species occurs. The species is utilized for spices and medicinal purposes, by using the seed and its aril. In contrast with the relatively narrow distribution of M. argentea, Myristica fatua Houtt. 1774 presents a large distribution, from the Malesian Archipelago to the Pacific islands (Lim Citation2012). Their distribution in these archipelagos makes them compelling subjects for investigating the biogeography of island plants in the region. Since their evolutionary history remains largely undocumented, the reconstruction of their chloroplast (cp) genome will help conducting phylogeographic studies.
Materials
Leaf samples of M. fatua originated from Tafea, Vanuatu (19° 35′ 52.8″ S, 169° 20′ 45.6″ E) were collected from New York Botanical Garden herbaria. A specimen was deposited at New York Botanical Garden (https://sweetgum.nybg.org/science/vh/specimen-details/?irn=3167800; Gregory M. Plunkett, [email protected]) under the voucher number 03509312. Further, leaf samples of M. argentea () from a silica-dried sample were collected in Ambon, Indonesia (3° 44′ 38.112″ S, 127° 55′ 47.568″ E). A specimen of M. argentea was deposited at UMR DIADE, IRD, France (https://dataverse.ird.fr/dataverse/umr_diade; [email protected]) under the number JK0155.
Figure 1. Myristica argentea grown in the forest of West Papua, Indonesia. (A) Tree trunk. (B) Mature fruit, with a description as follows: fruits solitary, ±ellipsoid-fusiform, 6.5–7.5 by 3.5–4.5 cm, apex narrowed, base somewhat contracted into a pseudostalk, 3–8 mm long, early glabrescent, brown, usually set with conspicuous coarse pale pustules or lenticels, similar as reported by de Wilde (Citation2014). (C) Longitudinal cut of the fruit showing red mace and nut (C). Pictures by Jakty Kusuma, Ambon, Indonesia, 2018. For the figure of Myristica fatua, picture is available in https://sweetgum.nybg.org/images3/3009/671/03509312.jpg.

Methods
Genomic DNA was extracted using the MATAB protocol following Mariac et al. (Citation2006). We constructed genomic libraries following protocol described in Mariac et al. (Citation2014). Libraries generated from real-time PCR were then sequenced on an Illumina NovaSeq (Illumina, San Diego, CA). Paired-end reads were filtered as described in Scarcelli et al. (Citation2016), and aligned to verified M. fragrans whole cp genome (OL628762). Cleaned reads were then assembled using the de novo assembler GetOrganelle ver.1.7.6.1 (Jin et al. Citation2020) (Supplementary Methods S1), and annotated using CPGAVAS2 (http://www.herbalgenomics.org/cpgavas2/) (Shi et al. Citation2019). We visualized the circular genome using CHLOROPLOT (https://irscope.shinyapps.io/Chloroplot/) (Zheng et al. Citation2020). All genomic information was deposited in GenBank (see Data availability statement).
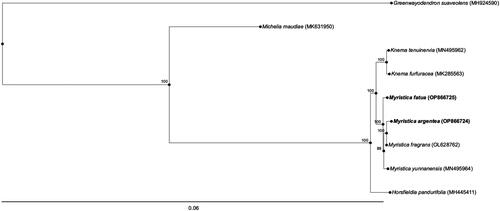
We then constructed a phylogenetic tree including the complete cp genome sequences of these two species as well as seven available complete cp genome sequences of closely related species of the family Myristicaceae (M. fragrans OL628762 (Cai et al. Citation2021); M. yunnanensis MN495964 (Mao et al. Citation2019); Knema tenuinervia MN495962 (Cai et al. Citation2021); Knema furfuracea MK285563 (Yang et al. Citation2019); and Horsfieldia amygdalina MK285561 (Zhang et al. Citation2019)), a species from Magnoliaceae (Michelia maudiae MK631950 (Wang et al. Citation2019)), and from Annonaceae, Greenwayodendron suaveolens MH924590 (Migliore et al. Citation2018) as an outgroup. MAFFT (https://mafft.cbrc.jp/alignment/server/index.html) ver. 7 (Katoh et al. Citation2019) and RAxML ver. 7.2.7 (https://cme.h-its.org/exelixis/php/countSource727.php) (Stamatakis Citation2014) were used to align the whole cpDNA sequences and infer a phylogenetic tree (Supplementary Methods S2).
Results
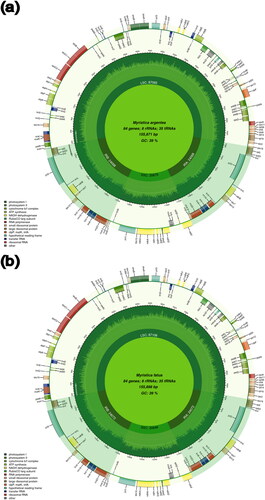
The full-length cp genome of M. argentea is 155,871 bp, while M. fatua is 155,898 bp and they exhibit respective assembly coverages of 41.2× and 31.8× ( and Supplementary Figure S1). The cpDNA genomes include a large single-copy (LSC) region (87,065 bp and 87,108 bp), a small single-copy (SSC) region (20,670 bp and 20,646 bp), and a pair of inverted repeat (IR) region (24,068 bp and 24,072 bp) in M. argentea and M. fatua, respectively. For both species, we identified 126 genes corresponding to 83 protein-coding genes, 35 tRNA, and eight rRNA, less genes in comparison with M. fragrans (146 genes) (de Oliveira et al. Citation2020), and slightly higher compared with M. yunnanensis (120 genes) (Yang et al. Citation2019).
Figure 2. Annotation of the M. argentea (a) and M. fatua (b) chloroplast genomes inferred by CPGAVAS2. The outer circle represents the genes annotated, classified by color based on their function. The length of genome is presented in the inner circle. Large single-copy (LSC), small single-copy (SSC), and inverted repeat regions (IRA and IRB) are marked.
Phylogenetic relations according to cpDNA genomes between species were well resolved (bootstrap support >85) (). Four Myristica species formed a monophyletic clade, showing a similar topology as previously reported (de Oliveira et al. Citation2020).
Figure 3. The best maximum-likelihood phylogenetic tree reconstructed using RAxML ver. 7.2.7 based on complete chloroplast genomes. The following sequences were used: M. fragrans (OL628762 (Cai et al. Citation2021)), M. yunnanensis (MN495964 (Mao et al. Citation2019)), Knema tenuinervia (MN495962 (Cai et al. Citation2021)), Knema furfuracea (MK285563 (Yang et al. Citation2019)), Horsfieldia pandurifolia (MH445411 (Zhang et al. Citation2019)), as well as Michelia maudiae (MK631950 (Wang et al. Citation2019)) and Greenwayodendron suaveolens (MH924590 (Migliore et al. Citation2018)) as outgroups. Numbers on branches indicate bootstrap values.
Discussion and conclusions
The cp genome structures of M. argentea and M. fatua have been analyzed and found to exhibit typical structural characteristics of angiosperm cp genomes, including one LSC and SSC region, as well as two IR regions. We identified a different length of atpF, clpP, and ndhD, genes in both species, with a difference from 1 to 3 bp. The cpDNA-based phylogenetic tree supports M. argentea as a sister species to M. fragrans, and the close taxonomic relationship between genera Knema and Myristica. This study will underpin the relationships between Myristica species and provide important details for evolutionary research and intra-specific diversity analysis.
Author contributions
JK and JD conducted the field work; JK, NS, and JD conceptualized and designed research; JK and MC performed lab experiment; JK and NS analyzed data; JK wrote original draft of the manuscript; all authors read and approved the final manuscript.
Ethical approval
This research was done with the authorization of Indonesian National Research and Innovation Agency (BRIN) with research permit to JD (144/SIP/FRP/E5/Ditk.KI/V/2018) and collecting permit to JK (B-3382/IPH.1/KS.02.04/IX/2019 and B-75/IV/KS.01.04/3/2022).
Supplemental Material
Download MS Word (99.5 KB)Supplemental Material
Download MS Word (38 KB)Acknowledgements
We thank Gregory M. Plunkett, Sean Thackurdeen, and Michael J. Balick from the New York Botanical Garden (NYBG) for having provided samples of M. fatua.
Disclosure statement
The authors report there are no competing interests to declare.
Data availability statement
The genome sequence data on this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) indicated in Bioproject (PRJNA904151), BioSample (M. argentea: SAMN31831556; M. fatua: SAMN31831555), and SRA (M. argentea: SRR22428069; M. fatua: SRR22428070). The species are registered under the accession numbers OP866724 (M. argentea) and OP866725 (M. fatua).
Additional information
Funding
References
- Cai C‐N, Ma H, Ci X‐Q, Conran JG, Li J. 2021. Comparative phylogenetic analyses of Chinese Horsfieldia (Myristicaceae) using complete chloroplast genome sequences. J Syst Evol. 59(3):504–514. doi: 10.1111/jse.12556.
- de Oliveira S, Duijm E, ter Steege H. 2020. Chloroplast genome of the nutmeg tree: Myristica fragrans Houtt. (Myristicaceae). bioRxiv.
- de Wilde WJJO. 2014. Flora Malesiana. 1st ed. Leiden, The Netherlands: Pensoft Publishers; p. 360–362.
- Govaerts R, Nic Lughadha E, Black N, Turner R, Paton A. 2021. The world checklist of vascular plants, a continuously updated resource for exploring global plant diversity. Sci Data. 8(1):215. doi: 10.1038/s41597-021-00997-6.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166. doi: 10.1093/bib/bbx108.
- Lim T. 2012. Myristica fatua. In: Edible medicinal and non-medicinal plants. 1st ed. Netherlands: Springer; p. 572–574. doi: 10.1007/978-94-007-2534-8.
- Mao C-L, Zhang F-L, Yang T, Li X-Q, Wu Y. 2019. The complete chloroplast genome sequence of Myristica yunnanensis (Myristicaceae). Mitochondrial DNA Part B. 4(1):1871–1872. doi: 10.1080/23802359.2019.1612711.
- Mariac C, Luong V, Kapran I, Mamadou A, Sagnard F, Deu M, Chantereau J, Gerard B, Ndjeunga J, Bezançon G, et al. 2006. Diversity of wild and cultivated pearl millet accessions (Pennisetum glaucum [L.] R. Br.) in Niger assessed by microsatellite markers. Theor Appl Genet. 114(1):49–58. doi: 10.1007/s00122-006-0409-9.
- Mariac C, Scarcelli N, Pouzadou J, Barnaud A, Billot C, Faye A, Kougbeadjo A, Maillol V, Martin G, Sabot F, et al. 2014. Cost-effective enrichment hybridization capture of chloroplast genomes at deep multiplexing levels for population genetics and phylogeography studies. Mol Ecol Resour. 14(6):1103–1113. doi: 10.1111/1755-0998.12258.
- Migliore J, Kaymak E, Mariac C, Couvreur TLP, Lissambou B‐J, Piñeiro R, Hardy OJ. 2018. Pre-Pleistocene origin of phylogeographical breaks in African rain forest trees: new insights from Greenwayodendron (Annonaceae) phylogenomics. J Biogeogr. 46:212–223. doi: 10.1111/jbi.13476.
- Scarcelli N, Mariac C, Couvreur TLP, Faye A, Richard D, Sabot F, Berthouly‐Salazar C, Vigouroux Y. 2016. Intra-individual polymorphism in chloroplasts from NGS data: where does it come from and how to handle it? Mol Ecol Resour. 16(2):434–445. [Database] doi: 10.1111/1755-0998.12462.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Wang J, Li Y, Wang Q, Fan W. 2019. Characterization of the complete chloroplast genome of Michelia maudiae (Magnoliaceae). Mitochondrial DNA B Resour. 4(2):2146–2147. doi: 10.1080/23802359.2019.1623110.
- Yang T, Mao C-L, Li X-Q, Zhang F-L, Zhao Q, Wu Y. 2019. The complete chloroplast genome sequence of Knema furfuracea (Myristicaceae). Mitochondrial DNA B Resour. 5(1):325–326. doi: 10.1080/23802359.2019.1703570.
- Zhang F-L, Mao C-L, Li X-Q, Yang T, Wu Y. 2019. The complete chloroplast genome sequence of Horsfieldia amygdalina (Myristicaceae). Mitochondrial DNA B Resour. 4(2):3923–3924. doi: 10.1080/23802359.2019.1688122.
- Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A. 2020. CHLOROPLOT: an online program for the versatile plotting of organelle genomes. Front. Genet. 11:576124. doi: 10.3389/fgene.2020.576124.
